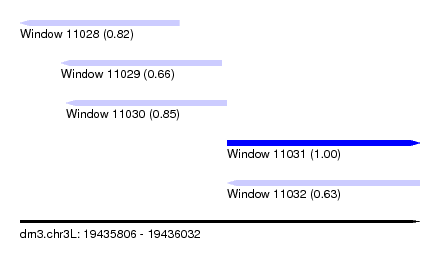
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,435,806 – 19,436,032 |
| Length | 226 |
| Max. P | 0.995369 |

| Location | 19,435,806 – 19,435,896 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 67.83 |
| Shannon entropy | 0.63460 |
| G+C content | 0.42918 |
| Mean single sequence MFE | -10.96 |
| Consensus MFE | -5.14 |
| Energy contribution | -5.39 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.816946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19435806 90 - 24543557 CCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCUCCUCUUUCCUCAAGCUCCAAC----------- ...(((((.......((..(((((......))))).....(((((...)))))..))...(((...........))).))))).......----------- ( -11.90, z-score = -1.03, R) >droEre2.scaffold_4784 19242528 90 - 25762168 CCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUCCCUCCUCCUUCCCCAACUUCCAGC----------- ...............((..(((((......))))).....(((((...)))))..)).................................----------- ( -9.10, z-score = -0.36, R) >droYak2.chr3L 3077802 90 + 24197627 CCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCUCCUCUUUCCCCAAGCUCCAAC----------- ...((((........((..(((((......))))).....(((((...)))))..))...(((...........)))..)))).......----------- ( -10.80, z-score = -0.85, R) >droSim1.chr3L 18764910 90 - 22553184 CCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCACCUCUUUCCCCAAGCUCCAAC----------- ...((((........((..(((((......))))).....(((((...)))))..))...(((...........)))..)))).......----------- ( -10.80, z-score = -0.93, R) >droMoj3.scaffold_6680 10404950 71 + 24764193 CCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCGCAAACACGAAUCCCAAUGCCAGC------------------------------ ....(((......(((((.(((((......)))))....)))))....)))....................------------------------------ ( -8.00, z-score = -0.25, R) >droGri2.scaffold_15110 23703654 76 - 24565398 CCACUUGACUAAACAGCACUCAAAAUUUAUUUUGAUUUAUGCUGCCCACAAGUACGGGAAAUAUCUCGAAUCGGAA------------------------- (((((((......(((((.((((((....))))))....)))))....))))).(((((....)))))....))..------------------------- ( -17.70, z-score = -3.24, R) >droWil1.scaffold_180727 730163 101 - 2741493 GCACUUGACUAAACAGAACUCAAAUUUGAUUUUGAUUUAUGCUACCCAUCCCAGCAUAGAGAUGUUUUAUCCCAUUUACUCCUUUCACCCAUUUACUAUCC ......((..(((((....(((((......)))))((((((((.........))))))))..)))))..)).............................. ( -12.20, z-score = -2.20, R) >droPer1.super_48 299005 77 - 581423 CCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUCUGCUGCUG--CUUCUCCAUUCUCCCUCUCUUUGUC---------------------- .............(((((.(((((......))))).....((...)))))))--.........................---------------------- ( -7.20, z-score = -0.27, R) >consensus CCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUCCACCUCUUUCCCCAA__UCCA_C___________ .............(((((.(((((......)))))....)))))......................................................... ( -5.14 = -5.39 + 0.25)

| Location | 19,435,829 – 19,435,920 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 71.06 |
| Shannon entropy | 0.56577 |
| G+C content | 0.42490 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -6.07 |
| Energy contribution | -6.07 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.661573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19435829 91 - 24543557 ---UAUCCCCACUCCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUC----------- ---.........((..(((..((((((....))))))...(((((.(((((......)))))....)))))...........)))..)).....----------- ( -14.80, z-score = -1.91, R) >droAna3.scaffold_13337 5253875 88 + 23293914 ------CAUUCUCCCCCCCAAAGUCAACCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACGAGACUGC----------- ------...............((((((....))))))...(((((.(((((......)))))....))))).....((((((....).))))).----------- ( -18.30, z-score = -3.47, R) >droEre2.scaffold_4784 19242551 86 - 25762168 -------ACCA-UCCUUCCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUCC----------- -------....-.........((((((....)))))).....((..(((((......))))).....(((((...)))))..))..........----------- ( -13.60, z-score = -1.47, R) >droYak2.chr3L 3077825 92 + 24197627 --CCAUCACCAUUCCUUCCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUC----------- --........((((.......((((((....)))))).....((..(((((......))))).....(((((...)))))..))...))))...----------- ( -14.40, z-score = -1.58, R) >droSec1.super_29 137513 91 - 700605 ---CAUUCCCACUCCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACCGAAUUUC----------- ---.........((..(((..((((((....))))))...(((((.(((((......)))))....)))))...........)))..)).....----------- ( -14.80, z-score = -2.17, R) >droSim1.chr3L 18764933 91 - 22553184 ---CAUCCCCACUCCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUC----------- ---.........((..(((..((((((....))))))...(((((.(((((......)))))....)))))...........)))..)).....----------- ( -14.80, z-score = -1.73, R) >droVir3.scaffold_13049 13019721 72 - 25233164 --------------CUAGCCUUG-CAGCCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCGCAAACACGAUAA------------------ --------------...((...(-((((....((.((....)))).(((((......))))).....)))))..))...........------------------ ( -11.10, z-score = -0.81, R) >droMoj3.scaffold_6680 10404962 71 + 24764193 --------------CUAGCCUUG-CAGCCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCGCAAACACGAAU------------------- --------------...((...(-((((....((.((....)))).(((((......))))).....)))))..))..........------------------- ( -11.10, z-score = -0.81, R) >droGri2.scaffold_15110 23703666 76 - 24565398 --------------CUAGCCUUG-UAGCCACUUGACUAAACAGCACUCAAAAUUUAUUUUGAUUUAUGCUGCCCACAAGUACGGGAAAUAU-------------- --------------.....((((-(....(((((......(((((.((((((....))))))....)))))....))))))))))......-------------- ( -13.70, z-score = -0.91, R) >droWil1.scaffold_180727 730186 105 - 2741493 AUGUUUCCGGUUUCGUGGCUGGCUCAGGCACUUGACUAAACAGAACUCAAAUUUGAUUUUGAUUUAUGCUACCCAUCCCAGCAUAGAGAUGUUUUAUCCCAUUUA ..((((..((((..(((.(((...))).)))..))))))))((((((((((......)))))((((((((.........))))))))...))))).......... ( -21.90, z-score = -0.62, R) >consensus ______CCCCA_UCCUUGCAAAGUCAGCCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAUUUC___________ ........................................(((((.(((((......)))))....))))).................................. ( -6.07 = -6.07 + -0.00)

| Location | 19,435,832 – 19,435,923 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 75.59 |
| Shannon entropy | 0.49071 |
| G+C content | 0.42390 |
| Mean single sequence MFE | -13.92 |
| Consensus MFE | -7.11 |
| Energy contribution | -6.87 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19435832 91 - 24543557 -AUCUAUCCCCACUCCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAU -............((..(((..((((((....))))))...(((((.(((((......)))))....)))))...........)))..)).. ( -14.80, z-score = -1.89, R) >droAna3.scaffold_13337 5253878 88 + 23293914 ----AACCAUUCUCCCCCCCAAAGUCAACCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACGAGAC ----......((((........((((((....))))))...(((((.(((((......)))))....)))))...............)))). ( -16.30, z-score = -3.44, R) >droEre2.scaffold_4784 19242554 86 - 25762168 -----AUCACCA-UCCUUCCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAU -----.((....-.........((((((....)))))).....((..(((((......))))).....(((((...)))))..))...)).. ( -14.00, z-score = -1.45, R) >droYak2.chr3L 3077828 92 + 24197627 AUCCCAUCACCAUUCCUUCCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAU ...........((((.......((((((....)))))).....((..(((((......))))).....(((((...)))))..))...)))) ( -14.30, z-score = -1.72, R) >droSec1.super_29 137516 91 - 700605 -AUCCAUUCCCACUCCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAACAGUCCGCACCGAAU -............((..(((..((((((....))))))...(((((.(((((......)))))....)))))...........)))..)).. ( -14.80, z-score = -2.40, R) >droSim1.chr3L 18764936 91 - 22553184 -AUUCAUCCCCACUCCUUGCAAAGUCAACCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAU -((((.................((((((....)))))).....((..(((((......))))).....(((((...)))))..))...)))) ( -15.20, z-score = -1.95, R) >droVir3.scaffold_13049 13019721 72 - 25233164 ---------------CUAGCCUUG-CAGCCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCGCAAACACGAUAA---- ---------------...((...(-((((....((.((....)))).(((((......))))).....)))))..))...........---- ( -11.10, z-score = -0.81, R) >droMoj3.scaffold_6680 10404962 71 + 24764193 ---------------CUAGCCUUG-CAGCCACUUGACUAAACAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCGCAAACACGAAU----- ---------------...((...(-((((....((.((....)))).(((((......))))).....)))))..))..........----- ( -11.10, z-score = -0.81, R) >droGri2.scaffold_15110 23703666 76 - 24565398 ---------------CUAGCCUUG-UAGCCACUUGACUAAACAGCACUCAAAAUUUAUUUUGAUUUAUGCUGCCCACAAGUACGGGAAAUAU ---------------.....((((-(....(((((......(((((.((((((....))))))....)))))....))))))))))...... ( -13.70, z-score = -0.91, R) >consensus _____AUC_CCA_UCCUUGCAAAGUCAGCCACUUGACUAAAUAGCACUCAAAUUUGAUUUUGAUUUAUGCUGCCCAGCAGUCCGCACCGAAU .........................................(((((.(((((......)))))....))))).................... ( -7.11 = -6.87 + -0.25)
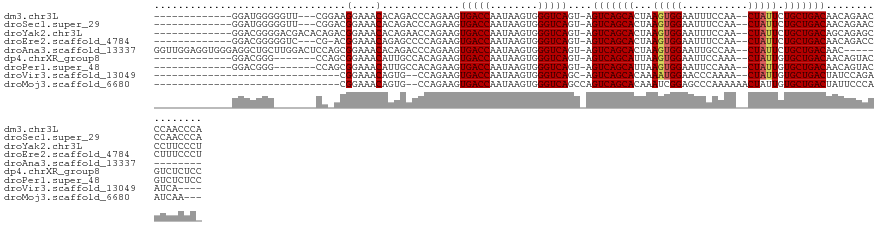
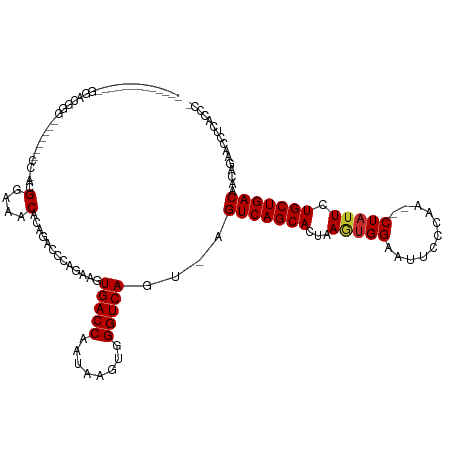
| Location | 19,435,923 – 19,436,032 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 128 |
| Reading direction | forward |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.53841 |
| G+C content | 0.49115 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.48 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.995369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19435923 109 + 24543557 -------------GGAUGGGGGUU---CGGAAGGAAACACAGACCCAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCACUAAGUGGAAUUUCCAA--CUAUUCUGCUGACAACAGAACCCAACCCA -------------((....(((((---(....(....)...((((((...((.....))...)))))).((-.(((((((...(((((.....)).)--))....))))))).)).))))))...)). ( -38.70, z-score = -2.81, R) >droSec1.super_29 137607 109 + 700605 -------------GGAUGGGGGUU---CGGACGGAAACACAGACCCAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCACUAAGUGGAAUUUCCAA--CUAUUCUGCUGACAACAGAACCCAACCCA -------------((....(((((---(....(....)...((((((...((.....))...)))))).((-.(((((((...(((((.....)).)--))....))))))).)).))))))...)). ( -38.80, z-score = -2.74, R) >droYak2.chr3L 3077920 112 - 24197627 -------------GGACGGGGACGACACAGACGGAAACACAGAACCAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCACUAAGUGGAAUUUCCAA--CUAUUCUGCUGACAGCAGAGCCCUUCCCU -------------((.((....))........(....)......)).(((((((((........)))))((-.(((((((...(((((.....)).)--))....))))))).))......))))... ( -32.50, z-score = -1.22, R) >droEre2.scaffold_4784 19242640 108 + 25762168 -------------GGACGGGGGUC---CG-ACGGAAACAGAGCCCCAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCACUAAGUGGAAUUUCCAA--CUAUUCUGCUGACAACAGACCCUUUCCCU -------------(((..((((..---(.-..(....).)..)))).................(((((.((-.(((((((...(((((.....)).)--))....))))))).)).)))))..))).. ( -38.90, z-score = -2.45, R) >droAna3.scaffold_13337 5253975 112 - 23293914 GGUUGGAGGUGGGAGGCUGCUUGGACUCCAGCGGAAACACAGACCCAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCACUAAGUGGAAUUGCCAA--CUAUUCUGCUGACAAC------------- .(((((((....(((....)))...)))))))(....)...((((((...((.....))...))))))...-.(((((((...(((((.....)).)--))....)))))))...------------- ( -36.10, z-score = -0.81, R) >dp4.chrXR_group8 7521063 105 + 9212921 -------------GGACGGG-------CCAGCGGAAACAUUGCCACAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCAUUAAGUGGAAUUCCAAA--CUAUUGUGCUGACAACAGUACGUCUCUCC -------------(((((((-------(.((.(....).)))))((.....(((((........)))))((-.((((((((....(((....)))..--.....)))))))).)).)).))))).... ( -35.10, z-score = -2.78, R) >droPer1.super_48 299114 105 + 581423 -------------GGACGGG-------CCAGCGGAAACAUUGCCACAGAAGUGACCAAUAAGUGGGUCAGU-AGUCAGCAUUAAGUGGAAUUCCAAA--CUAUUGUGCUGACAACAGUACGUCUCUCC -------------(((((((-------(.((.(....).)))))((.....(((((........)))))((-.((((((((....(((....)))..--.....)))))))).)).)).))))).... ( -35.10, z-score = -2.78, R) >droVir3.scaffold_13049 13019793 88 + 25233164 -------------------------------CGGAAACAGUG--CCAGAAGUGACCAAUAAGUGGGUCAGC-AGUCAGCACAAAAUGGAACCCAAAA--CUAUUGUGCUGACUAUCCAGAAUCA---- -------------------------------.(((.......--.......(((((........)))))..-((((((((((..((((.........--)))))))))))))).))).......---- ( -26.30, z-score = -2.73, R) >droMoj3.scaffold_6680 10405033 92 - 24764193 -------------------------------CGGAAACAGUG--CCAGAAGUGACCAAUAAGUGGGUCAGCCAGUCAGCACAAAUCGGAGCCCAAAAAACUAUUGUGCUGACUAUUCCCAAUCAA--- -------------------------------.((((......--.......(((((........)))))...(((((((((((...................))))))))))).)))).......--- ( -25.21, z-score = -2.23, R) >consensus _____________GGACGGG_______CC_ACGGAAACACAGACCCAGAAGUGACCAAUAAGUGGGUCAGU_AGUCAGCACUAAGUGGAAUUCCCAA__CUAUUCUGCUGACAACAGAACCUCACCC_ ................................(....).............(((((........)))))....(((((((...(((((...........))))).)))))))................ (-19.34 = -19.48 + 0.14)
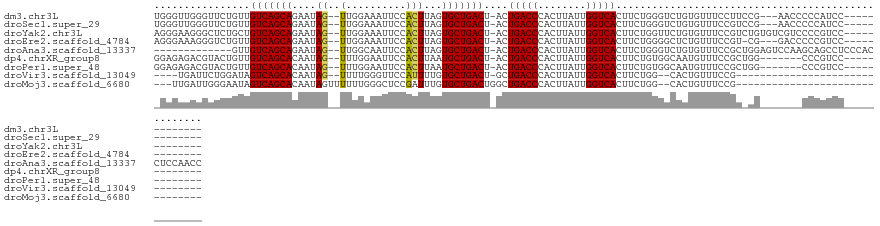
| Location | 19,435,923 – 19,436,032 |
|---|---|
| Length | 109 |
| Sequences | 9 |
| Columns | 128 |
| Reading direction | reverse |
| Mean pairwise identity | 71.71 |
| Shannon entropy | 0.53841 |
| G+C content | 0.49115 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -13.82 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.630418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19435923 109 - 24543557 UGGGUUGGGUUCUGUUGUCAGCAGAAUAG--UUGGAAAUUCCACUUAGUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGGGUCUGUGUUUCCUUCCG---AACCCCCAUCC------------- .((((.((((((....(((((((...(((--.(((.....))).))).)))))))(-((.((((((...............)))))).))).........)---)))))..))))------------- ( -36.36, z-score = -2.12, R) >droSec1.super_29 137607 109 - 700605 UGGGUUGGGUUCUGUUGUCAGCAGAAUAG--UUGGAAAUUCCACUUAGUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGGGUCUGUGUUUCCGUCCG---AACCCCCAUCC------------- .((((.((((((....(((((((...(((--.(((.....))).))).)))))))(-((.((((((...............)))))).))).........)---)))))..))))------------- ( -36.36, z-score = -1.95, R) >droYak2.chr3L 3077920 112 + 24197627 AGGGAAGGGCUCUGCUGUCAGCAGAAUAG--UUGGAAAUUCCACUUAGUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGGUUCUGUGUUUCCGUCUGUGUCGUCCCCGUCC------------- .((((..((((((((.....)))))((((--.(((((((.(((..((((....)))-).(((((........)))))....)))......))))))).))))))).)))).....------------- ( -32.80, z-score = -0.69, R) >droEre2.scaffold_4784 19242640 108 - 25762168 AGGGAAAGGGUCUGUUGUCAGCAGAAUAG--UUGGAAAUUCCACUUAGUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGGGGCUCUGUUUCCGU-CG---GACCCCCGUCC------------- ..(((..(((((.((.(((((((...(((--.(((.....))).))).))))))).-)).))))).................((((.((((.......-))---)).)))).)))------------- ( -40.80, z-score = -2.33, R) >droAna3.scaffold_13337 5253975 112 + 23293914 -------------GUUGUCAGCAGAAUAG--UUGGCAAUUCCACUUAGUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGGGUCUGUGUUUCCGCUGGAGUCCAAGCAGCCUCCCACCUCCAACC -------------((.(((((((...(((--.(((.....))).))).))))))).-))(((((........)))))....((((.((((..((((...)))).....))))...))))......... ( -30.20, z-score = -0.42, R) >dp4.chrXR_group8 7521063 105 - 9212921 GGAGAGACGUACUGUUGUCAGCACAAUAG--UUUGGAAUUCCACUUAAUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGUGGCAAUGUUUCCGCUGG-------CCCGUCC------------- .....((((..((((.(((((((....((--(..((....)))))...))))))).-))(((((........))))).....((((........)))).))-------..)))).------------- ( -28.10, z-score = -0.80, R) >droPer1.super_48 299114 105 - 581423 GGAGAGACGUACUGUUGUCAGCACAAUAG--UUUGGAAUUCCACUUAAUGCUGACU-ACUGACCCACUUAUUGGUCACUUCUGUGGCAAUGUUUCCGCUGG-------CCCGUCC------------- .....((((..((((.(((((((....((--(..((....)))))...))))))).-))(((((........))))).....((((........)))).))-------..)))).------------- ( -28.10, z-score = -0.80, R) >droVir3.scaffold_13049 13019793 88 - 25233164 ----UGAUUCUGGAUAGUCAGCACAAUAG--UUUUGGGUUCCAUUUUGUGCUGACU-GCUGACCCACUUAUUGGUCACUUCUGG--CACUGUUUCCG------------------------------- ----.......(((((((((((((((...--...(((...)))..)))))))))))-).(((((........))))).......--.......))).------------------------------- ( -26.70, z-score = -2.30, R) >droMoj3.scaffold_6680 10405033 92 + 24764193 ---UUGAUUGGGAAUAGUCAGCACAAUAGUUUUUUGGGCUCCGAUUUGUGCUGACUGGCUGACCCACUUAUUGGUCACUUCUGG--CACUGUUUCCG------------------------------- ---......(((((((((((((((((.((((.....)))).....)))))))))(..(.(((((........)))))...)..)--..)))))))).------------------------------- ( -29.50, z-score = -2.44, R) >consensus _GGGAGAGGUUCUGUUGUCAGCAGAAUAG__UUGGAAAUUCCACUUAGUGCUGACU_ACUGACCCACUUAUUGGUCACUUCUGGGUCUGUGUUUCCGU_CG_______CCCGUCC_____________ ................(((((((.........................)))))))....(((((........)))))................................................... (-13.82 = -13.82 + 0.00)
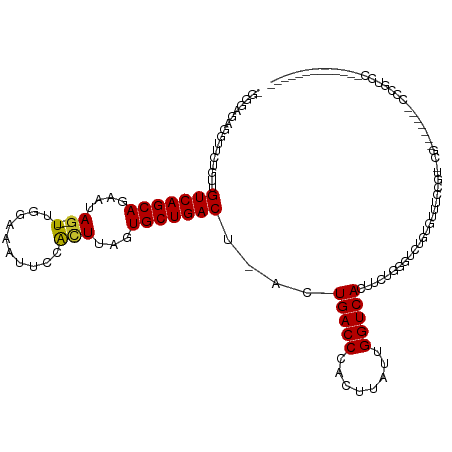
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:48 2011