
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,435,638 – 19,435,744 |
| Length | 106 |
| Max. P | 0.909487 |

| Location | 19,435,638 – 19,435,744 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.66 |
| Shannon entropy | 0.51198 |
| G+C content | 0.44701 |
| Mean single sequence MFE | -27.45 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.01 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19435638 106 - 24543557 GGCUCAUAACUUUGGCAUCUAUCGCAGUAUG-------CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACUAACACUCAAAGAAA-- .............(((((((.....)).)))-------)).(((............(((((((((((......)))))))).)))....(((((.......)))))...))).-- ( -28.40, z-score = -2.56, R) >droAna3.scaffold_13337 5253656 108 + 23293914 GGCUCAUAACUUUGGCAUCUAUCGUCGUUUG-------CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUAGAAUUCUGGCAUUGAAAAAAA .((.((......))))........(((..((-------((.(((.((((((.((..(((((((((((......)))))))).))).)))))))))))....)))).)))...... ( -28.20, z-score = -1.89, R) >droEre2.scaffold_4784 19242366 106 - 25762168 GCCUCAUAACUUUGGCAUCUAGCGCAGUAUG-------CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAGACGGAGCGCUAUUUACACGCAGCGAAA-- (((..........)))...((((((((((((-------.....))))))..(...((((((((((((......)))))))).))))...).))))))................-- ( -33.90, z-score = -2.17, R) >droYak2.chr3L 3077641 106 + 24197627 GGCUCAUAACUUUGGCAUCUAGCGUAGUAUG-------CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACAGAGAAGGAGUGCCAUUUACACGCAGCGAAA-- .((((.......((((.......((((((((-------.....)))).))))..(((((((((((((......))))))).......)))))))))).......).)))....-- ( -28.95, z-score = -0.89, R) >droSec1.super_29 137326 106 - 700605 GGCUCAUAACUUUGGCAUCCAUCGCAGUAUG-------CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACGUACACUCAGAGAAA-- ..(((........(((((.(......).)))-------))................(((((((((((......)))))))).)))....(((((.......))))).)))...-- ( -29.60, z-score = -1.89, R) >droSim1.chr3L 18764746 106 - 22553184 GGCUCAUAACUUUGGCAUCCAUCGCAGUAUG-------CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUCGGCGUGUAACGGAAACGGAGUGAAACGUACACUCAGAGAAA-- ..(((........(((((.(......).)))-------))................(((((((((((......)))))))).)))....(((((.......))))).)))...-- ( -29.60, z-score = -1.89, R) >droVir3.scaffold_13049 13019547 103 - 25233164 AUCUCAUAACUCUGGCAUCUAUCGGCCG-----------CUUUGGUAUUUACAUGCUGCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUAGCAUUGAGCACUCAUGAAAG- ...((((......(((........)))(-----------(((((.((((((....(((.((((((((......)))))))))))....)))))).))..))))....))))...- ( -30.50, z-score = -1.76, R) >droMoj3.scaffold_6680 10404801 104 + 24764193 AUCUCAUAACUCUGGCAUCUAUCGGCCG----------CCUCUCGUAUUUACAUGCUGCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUAGCAUUGAGCACUUGUGAAAG- ...(((((((((.(((........)))(----------(..(((((....))...(((.((((((((......))))))))))).....)))..))...)))...))))))...- ( -30.00, z-score = -1.37, R) >droGri2.scaffold_15110 23703476 102 - 24565398 AUCUUAUAACUUUGGCAUCUAUCUACCG-----------CUUUCAUAUUUACAUGCUGCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUUGAAUUGAGCUCUCAUGAAA-- .............(((...........)-----------))(((((.........(((.((((((((......))))))))))).....(((((......)))))..))))).-- ( -21.10, z-score = -0.58, R) >droWil1.scaffold_180727 730012 93 - 2741493 GGCUUAUAACAUUGGCAUCUAUCGUCUCGU--UUCG--CCUUUCAUAUUUACAUGCUUCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUAGGA------------------ (((....(((...(((.......)))..))--)..)--))..((.((((((.((..(((((((((((......)))))))).))).)))))))).))------------------ ( -20.50, z-score = -0.52, R) >dp4.chrXR_group8 7520776 99 - 9212921 GGCUCAUAACUUUGGCAUCUAUCGUCCUUUCAUUCGUUCCUUUCAUAUUUACAUGCUUCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGAAGAGCU---------------- (((((........(((.......))).(((((....((((...(((......)))....((((((((......)))))))).))))..))))).)))))---------------- ( -24.30, z-score = -2.00, R) >droPer1.super_48 298834 99 - 581423 GGCUCAUAACUUUGGCAUCUAUCGUCCUUUCAUUCGUUCCUUUCAUAUUUACAUGCUUCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGAAGAGCU---------------- (((((........(((.......))).(((((....((((...(((......)))....((((((((......)))))))).))))..))))).)))))---------------- ( -24.30, z-score = -2.00, R) >consensus GGCUCAUAACUUUGGCAUCUAUCGUCGUAUG_______CCUUUCAUAUUUACAUGCUCCUUACACGCGCAUUGGCGUGUAACGGAAAUUGAGUAGAAUUUACAC_CAGAGAAA__ ..(((.........((((.((............................)).))))(((((((((((......)))))))).)))....)))....................... (-14.77 = -15.01 + 0.24)

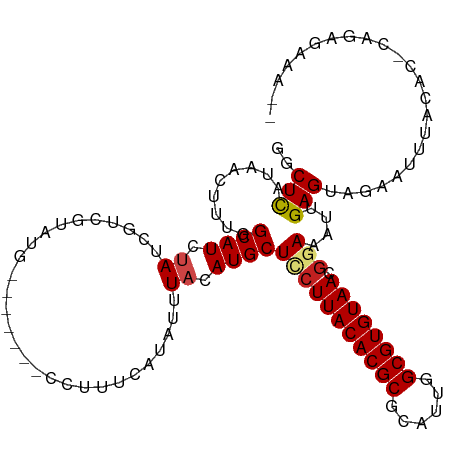
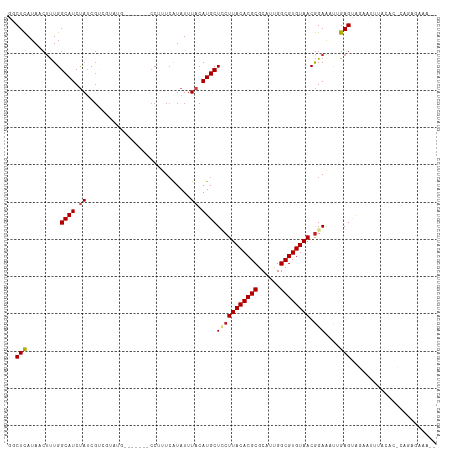
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:44 2011