| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,428,907 – 19,429,001 |
| Length | 94 |
| Max. P | 0.950583 |

| Location | 19,428,907 – 19,429,001 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.54 |
| Shannon entropy | 0.55379 |
| G+C content | 0.45979 |
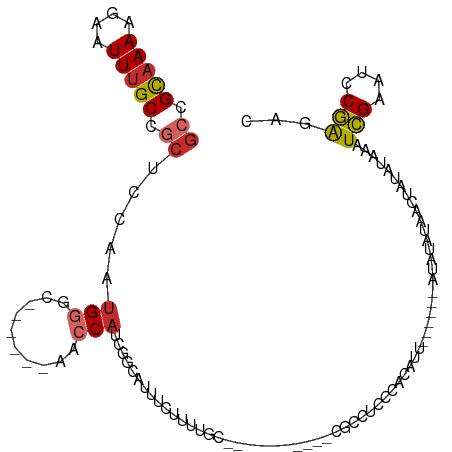
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -5.99 |
| Energy contribution | -6.04 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.30 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.950583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
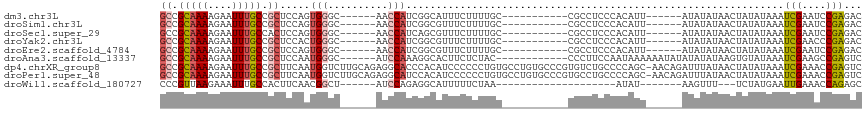
>dm3.chr3L 19428907 94 - 24543557 GCCGCAAAAGAAUUUGCCGCUCCAGUGGGC------AACCAUCGGCAUUUCUUUUGC-----------CGCCUCCCACAUU------AUAUAUAACUAUAUAAAUCGAAUCCGAGAC ((.(((((((((..(((((.....((((..------..))))))))).)))))))))-----------.))........((------(((((....))))))).(((....)))... ( -31.10, z-score = -5.38, R) >droSim1.chr3L 18758085 94 - 22553184 GCCGCAAAAGAAUUUGCCGCUCCAGUGGGC------AACCAUCGGCGUUUCUUUUGC-----------CGCCUCCCACAUU------AUAUAUAACUAUAUAAAUCGAAUCCGAGAC ((.(((((((((..(((((.....((((..------..))))))))).)))))))))-----------.))........((------(((((....))))))).(((....)))... ( -30.40, z-score = -4.76, R) >droSec1.super_29 130955 94 - 700605 GCCGCAAAAGAAUUUGCCACUCCAGUGGGC------AACCAUCAGCGUUUCUUUUGC-----------CGCCUCCCACAUU------AUAUAUAACUAUAUAAAUCGAAUCCGAGAC ((.(((((((((..(((.......((((..------..))))..))).)))))))))-----------.))........((------(((((....))))))).(((....)))... ( -23.60, z-score = -3.75, R) >droYak2.chr3L 3070756 94 + 24197627 GCCGCAAAAGAAUUUGCCGCUCCACUGGGC------AACCAUCGGCGUUUCUUUUGC-----------CGCCUCCCACAUU------AUAUAUAACUAUAUAAAUCGAACCCGAGAC ((.(((((((((..(((((......(((..------..))).))))).)))))))))-----------.))........((------(((((....))))))).(((....)))... ( -28.10, z-score = -4.66, R) >droEre2.scaffold_4784 19234380 94 - 25762168 GCCGCAAAAGAAUUUGCCGCUCCAGUGGGC------AACCAUCGGCGUUUCUUUUGC-----------CGCCUCCCACAUU------AUAUAUAACUAUAUAAAUCGAAUCCGAGAC ((.(((((((((..(((((.....((((..------..))))))))).)))))))))-----------.))........((------(((((....))))))).(((....)))... ( -30.40, z-score = -4.76, R) >droAna3.scaffold_13337 5247306 99 + 23293914 GCCGCAAAAGAAUUUGCCGCUCCAAUGGGC------AUCCAAAGGCACUUCUCUAC------------CCCUUCCAAUAAAAAAUAUAUAUAUAAGUGUAUAAAUCGAAGCCGAGUC ((((((((....))))).((((....))))------.......)))..........------------................(((((((....)))))))..(((....)))... ( -17.30, z-score = -0.97, R) >dp4.chrXR_group8 7514554 116 - 9212921 GCCGCAAAAGAAUUUGCCGCUUCAAUGGUCUUGCAGAGGCACCCACAUCCCCCCUGUGCCUGUGCCCGUGUCUGCCCCAGC-AACAGAUUUAUAACUAUAUAAAUCGAAACCGAGUC ((.(((((....))))).)).....((((.(((((.((((((.............)))))).)))...(((.(((....))-))))((((((((....)))))))))).)))).... ( -31.02, z-score = -3.10, R) >droPer1.super_48 292568 116 - 581423 GCCGCAAAAGAAUUUGCCGCUUCAAUGGUCUUGCAGAGGCAUCCACAUCCCCCCUGUGCCUGUGCCCGUGCCUGCCCCAGC-AACAGAUUUAUAACUAUAUAAAUCGAAACCGAGUC ((.(((((....))))).))(((...(((...(((.((((((.............)))))).)))....)))(((....))-)...((((((((....)))))))))))........ ( -28.82, z-score = -2.21, R) >droWil1.scaffold_180727 724945 81 - 2741493 CCCGUUAAGAAAUUUGCCACUUCAACGGCU------AUCCAGAGGCAUUUUUCUAA--------------------AUAU-------AAGUUU---UCUAUGAAUUGAAACCAGAGC ...(((.(((((..((((.((.....((..------..)))).))))..))))).)--------------------))..-------..(.((---((........)))).)..... ( -12.40, z-score = -0.30, R) >consensus GCCGCAAAAGAAUUUGCCGCUCCAAUGGGC______AACCAUCGGCAUUUCUUUUGC___________CGCCUCCCACAUU______AUAUAUAACUAUAUAAAUCGAAUCCGAGAC ((.(((((....))))).))....................................................................................(((....)))... ( -5.99 = -6.04 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:43 2011