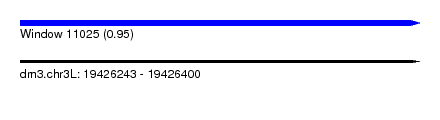
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,426,243 – 19,426,400 |
| Length | 157 |
| Max. P | 0.952338 |

| Location | 19,426,243 – 19,426,400 |
|---|---|
| Length | 157 |
| Sequences | 8 |
| Columns | 171 |
| Reading direction | forward |
| Mean pairwise identity | 65.14 |
| Shannon entropy | 0.66175 |
| G+C content | 0.42450 |
| Mean single sequence MFE | -46.01 |
| Consensus MFE | -11.65 |
| Energy contribution | -10.34 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.952338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
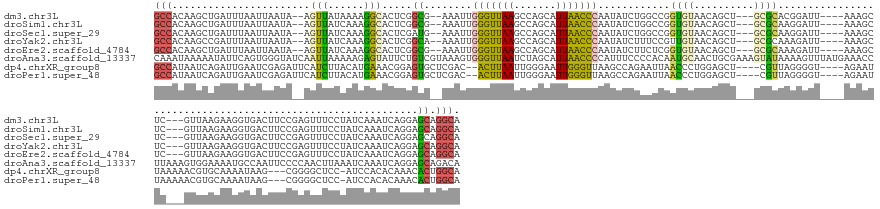
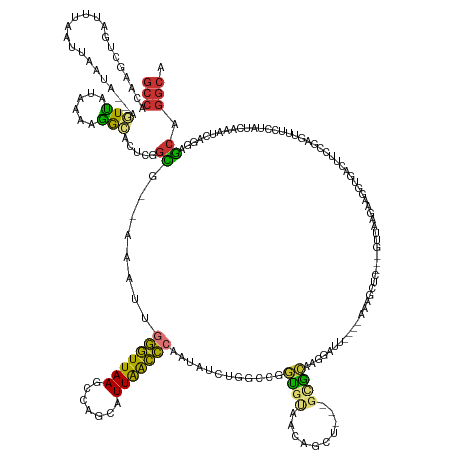
>dm3.chr3L 19426243 157 + 24543557 GCCACAAGCUGAUUUAAUUAAUA--AGUUAUAAAAGGCACUCGGCG--AAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCU---GCGCACGGAUU----AAAGCUC---GUUAAGAAGGUGACUUCCGAGUUUCCUAUCAAAUCAGGAGCAGGCA (((...(((((...(((((....--)))))......((((.(((((--(.((((((((((.......)))))))))).))..))))))))..)))))---..((.......----..(((((---(....((((....))))))))))((((........)))))).))). ( -54.60, z-score = -4.09, R) >droSim1.chr3L 18755424 157 + 22553184 GCCACAAGCUGAUUUAAUUAAUA--AGUUAUCAAAGGCACUCGGCG--AAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCU---GCGCAAGGAUU----AAAGCUC---GUUAAGAAGGUGACUUCCGAGUUUCCUAUCAAAUCAGGAGCAGGCA (((...(((((...(((((....--)))))......((((.(((((--(.((((((((((.......)))))))))).))..))))))))..)))))---..((.......----..(((((---(....((((....))))))))))((((........)))))).))). ( -54.60, z-score = -4.14, R) >droSec1.super_29 128278 157 + 700605 GCCACAAGCUGAUUUAAUUAAUA--AGUUAUCAAAGGCACUCGAUG--AAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCU---GCGCAAGGAUU----AAAGCUC---GUUAAGAAGGUGACUUCCGAGUUUCCUAUCAAAUCAGGAGCAGGCA (((...(((((...(((((....--)))))......(((((.(..(--(.((((((((((.......)))))))))).))...).)))))..)))))---..((.......----..(((((---(....((((....))))))))))((((........)))))).))). ( -49.30, z-score = -2.98, R) >droYak2.chr3L 3068080 157 - 24197627 GCCACAAGCCGAUUUAAUUAAUA--AGUUAUCAAAGGCACUCGGCA--AAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUUCCGUUGUAACAGCU---GCGCAAAGAUU----AAAGCUC---GUUAAGAAGGUGACUUCCGAGUUUCCUAUCAAAUCAGGAGCAGGCA (((....(((....(((((....--))))).....)))(((((((.--..((((((((((.......))))))))))((((((.(((.((....)).---))).)))))).----...))..---.....((((....)))))))))(((((........)))))..))). ( -47.60, z-score = -4.01, R) >droEre2.scaffold_4784 19231791 157 + 25762168 GCCACAAGCUGAUUUAAUUAAUA--AGUUAUCAAAGGCACUCGGCG--AAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUUCUCGGUGUAACAGCU---GCGCAAAGAUU----AAAGCUC---GUUAAGAAGGUGACUUCCGAGUUUCCUAUCAAAUCAGGAGCAGGCA (((...(((((...(((((....--)))))......(((((.((.(--(.((((((((((.......)))))))))).))..)).)))))..)))))---..((.......----..(((((---(....((((....))))))))))((((........)))))).))). ( -49.10, z-score = -3.60, R) >droAna3.scaffold_13337 5243616 171 - 23293914 CAAAUAAAAAUAUUCAGUGGGUAUCAAUUAAAAAGAGUAUUCUGUCGUAAAGUGGGUUAAUCUAGCAUUAACCCCAUUUCCCCACAAUGCAACUGCGAAAGUAUAAAAGUUUAUGAAACCUUAAAGUGGAAAAUGCCAAUUCCCCAACUUAAAUCAAAUCAGGAGCAGACA ................(((((......(((...((......))....))).(.((((((((.....))))))))).....)))))........(((....))).....((((.(((....((((..(((..(((....)))..)))..))))......))).))))..... ( -28.70, z-score = -0.17, R) >dp4.chrXR_group8 7511931 157 + 9212921 GCCAUAAUCAGAUUGAAUCGAGAUUCAUCUUACAUGAAACGGAGUGCUCGAC--ACUUAAUUGGGAAUUGGGUUAAGCCAGAAUUAACCCUGGAGCU----CGUUAGGGGU----AGAAUUAAAAACGUGCAAAAUAAG---CGGGGCUCC-AUCCACACAAACACUGGCA ((((......((((((.(((((((((.((......))....)))).))))).--..)))))).(((...(((((((.......)))))))(((((((----(.((((....----....))))...(((.........)---)))))))))-))))..........)))). ( -42.10, z-score = -1.43, R) >droPer1.super_48 289940 157 + 581423 GCCAUAAUCAGAUUGAAUCGAGAUUCAUCUUACAUGAAACGGAGUGCUCGAC--ACUUAAUUGGGAAUUGGGUUAAGCCAGAAUUAACCCUGGAGCU----CGUUAGGGGU----AGAAUUAAAAACGUGCAAAAUAAG---CGGGGCUCC-AUCCACACAAACACUGGCA ((((......((((((.(((((((((.((......))....)))).))))).--..)))))).(((...(((((((.......)))))))(((((((----(.((((....----....))))...(((.........)---)))))))))-))))..........)))). ( -42.10, z-score = -1.43, R) >consensus GCCACAAGCUGAUUUAAUUAAUA__AGUUAUAAAAGGCACUCGGCG__AAAUUGGGUUAAGCCAGCAUUAACCCAAUAUCUGGCCGGUGUAACAGCU___GCGCAAGGAUU____AAAGCUC___GUUAAGAAGGUGACUUCCGAGUUUCCUAUCAAAUCAGGAGCAGGCA (((.......................(((......))).....((........(((((((.......)))))))....................((......))............................................................)).))). (-11.65 = -10.34 + -1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:43 2011