
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,413,914 – 19,414,006 |
| Length | 92 |
| Max. P | 0.896394 |

| Location | 19,413,914 – 19,414,006 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.38787 |
| G+C content | 0.46916 |
| Mean single sequence MFE | -30.01 |
| Consensus MFE | -22.07 |
| Energy contribution | -23.09 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.896394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
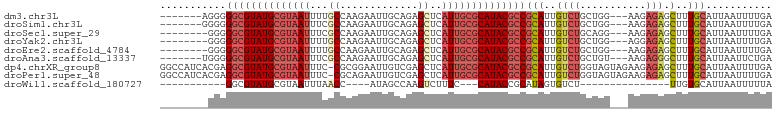
>dm3.chr3L 19413914 92 + 24543557 -------AGGGGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGG---AAGAGAGCUUUGCAUUAAUUUUGA -------....(((((((((((((((((((........)))))....))))))))))))))(((..((((.....---...))).)..)))........... ( -32.20, z-score = -1.78, R) >droSim1.chr3L 18739228 92 + 22553184 -------GGGGGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGG---AAGAGAGCUUUGCAUUAAUUUUGA -------....(((((((((((((((((((........)).)))...))))))))))))))(((..((((.....---...))).)..)))........... ( -30.20, z-score = -0.89, R) >droSec1.super_29 109549 91 + 700605 --------GGGGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCAGG---AAGAGAGCUUUGCAUUAAUUUUGA --------.(.(((((((((((((((((((........)).)))...)))))))))))))).)......((((((---........)))))).......... ( -30.30, z-score = -1.28, R) >droYak2.chr3L 3058118 91 - 24197627 --------GGGGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGG---AGGAGAGCUUUGCAUUAAUUUUGA --------...(((((((((((((((((((........)))))....))))))))))))))(((..((((.(...---.).))).)..)))........... ( -34.20, z-score = -2.36, R) >droEre2.scaffold_4784 19222140 91 + 25762168 --------GGGGGCGUAUGCGUAAUUUUGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGG---AAGAGAGCUUUGCAUUAAUUUUGA --------...(((((((((((((((((((........)))))....))))))))))))))(((..((((.....---...))).)..)))........... ( -32.20, z-score = -1.87, R) >droAna3.scaffold_13337 5234316 92 - 23293914 -------UGGGGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGU---AAGAGGGCUUUGCAUUAAUUCUGA -------....(((((((((((((((((((........)).)))...))))))))))))))(((..((((.(...---..).))))..)))........... ( -33.30, z-score = -1.72, R) >dp4.chrXR_group8 7502903 101 + 9212921 GGCCAUCACGAGGCGUAUGCGUAAUUUC-CGCGGAAUUGUCGAGCUCAUUGCGCAUACGCCGCAUUGUCUGGUAGUAGAAGAGAGCUUUGCAUUAAUUUUGA .((((..((((((((((((((((((...-..(((.....))).....))))))))))))))...)))).)))).(((((.......)))))........... ( -31.80, z-score = -0.92, R) >droPer1.super_48 280966 101 + 581423 GGCCAUCACGAGGCGUAUGCGUAAUUUC-CGCAGAAUUGUCGAGCUCAUUGCGCAUACGCCGCAUUGUCUGGUAGUAGAAGAGAGCUUUGCAUUAAUUUUGA .((((..(((((((((((((((((((((-.((......)).)))...))))))))))))))...)))).)))).(((((.......)))))........... ( -31.90, z-score = -1.32, R) >droWil1.scaffold_180727 697628 69 + 2741493 -----------GGCGUAUGCGUAAUUUAACC----AUAGCCAAGUCUUUC---CAUACCGCAUAGUGUCU---------------UUGUGCAUUAAUUUUUA -----------(((.((((.((......)))----)))))).........---......((((((.....---------------))))))........... ( -14.00, z-score = -1.80, R) >consensus ________GGGGGCGUAUGCGUAAUUUCGCCAAGAAUUGCAGAGCUCAUUGCGCAUACGCCGCAUUGUCUGCUGG___AAGAGAGCUUUGCAUUAAUUUUGA ...........((((((((((((((...((.............))..))))))))))))))(((..((((...........))).)..)))........... (-22.07 = -23.09 + 1.02)


| Location | 19,413,914 – 19,414,006 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 81.04 |
| Shannon entropy | 0.38787 |
| G+C content | 0.46916 |
| Mean single sequence MFE | -23.32 |
| Consensus MFE | -16.97 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
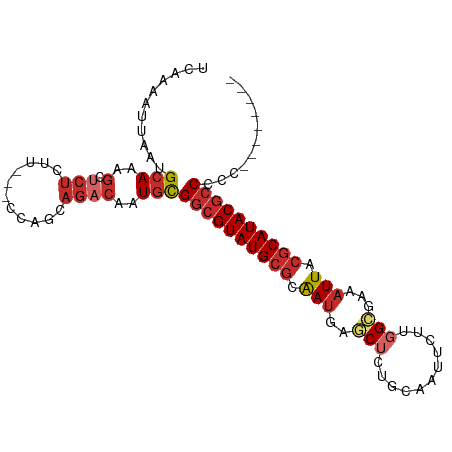
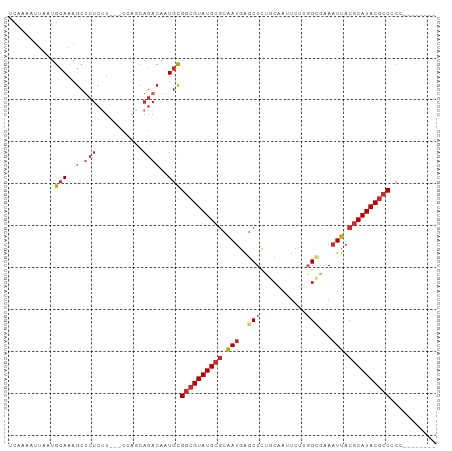
>dm3.chr3L 19413914 92 - 24543557 UCAAAAUUAAUGCAAAGCUCUCUU---CCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCCCCU------- ........................---...(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))....------- ( -24.10, z-score = -1.53, R) >droSim1.chr3L 18739228 92 - 22553184 UCAAAAUUAAUGCAAAGCUCUCUU---CCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCCCCC------- ........................---...(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))....------- ( -25.20, z-score = -1.73, R) >droSec1.super_29 109549 91 - 700605 UCAAAAUUAAUGCAAAGCUCUCUU---CCUGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCCCC-------- ...........(((..(.((((..---...).))))..)))((((((((((.(((..(((..(.....)..)))...))).))))))))))...-------- ( -25.40, z-score = -1.88, R) >droYak2.chr3L 3058118 91 + 24197627 UCAAAAUUAAUGCAAAGCUCUCCU---CCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCCCC-------- ...........(((..(.(((.(.---...).))))..)))((((((((((.(((..(((..(.....)..)))...))).))))))))))...-------- ( -24.30, z-score = -1.84, R) >droEre2.scaffold_4784 19222140 91 - 25762168 UCAAAAUUAAUGCAAAGCUCUCUU---CCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCAAAAUUACGCAUACGCCCCC-------- ........................---...(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))...-------- ( -24.10, z-score = -1.68, R) >droAna3.scaffold_13337 5234316 92 + 23293914 UCAGAAUUAAUGCAAAGCCCUCUU---ACAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCCCCA------- ........................---...(((.....)))((((((((((.(((..(((..(.....)..)))...))).))))))))))....------- ( -25.20, z-score = -1.30, R) >dp4.chrXR_group8 7502903 101 - 9212921 UCAAAAUUAAUGCAAAGCUCUCUUCUACUACCAGACAAUGCGGCGUAUGCGCAAUGAGCUCGACAAUUCCGCG-GAAAUUACGCAUACGCCUCGUGAUGGCC ..............................(((....(((.((((((((((.(((..((..((....)).)).-...))).)))))))))).)))..))).. ( -26.40, z-score = -1.24, R) >droPer1.super_48 280966 101 - 581423 UCAAAAUUAAUGCAAAGCUCUCUUCUACUACCAGACAAUGCGGCGUAUGCGCAAUGAGCUCGACAAUUCUGCG-GAAAUUACGCAUACGCCUCGUGAUGGCC ..............................(((....(((.((((((((((.(((...(.((.((....))))-)..))).)))))))))).)))..))).. ( -27.10, z-score = -1.55, R) >droWil1.scaffold_180727 697628 69 - 2741493 UAAAAAUUAAUGCACAA---------------AGACACUAUGCGGUAUG---GAAAGACUUGGCUAU----GGUUAAAUUACGCAUACGCC----------- .................---------------......((((((((.(.---...).))(((((...----.)))))....))))))....----------- ( -8.10, z-score = 0.79, R) >consensus UCAAAAUUAAUGCAAAGCUCUCUU___CCAGCAGACAAUGCGGCGUAUGCGCAAUGAGCUCUGCAAUUCUUGGCGAAAUUACGCAUACGCCCCC________ .........................................((((((((((.(((..((.............))...))).))))))))))........... (-16.97 = -17.50 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:40 2011