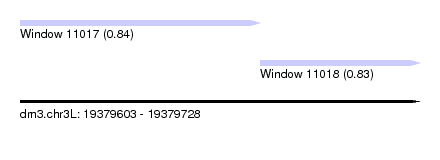
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,379,603 – 19,379,728 |
| Length | 125 |
| Max. P | 0.838299 |

| Location | 19,379,603 – 19,379,678 |
|---|---|
| Length | 75 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 60.92 |
| Shannon entropy | 0.61619 |
| G+C content | 0.30141 |
| Mean single sequence MFE | -16.13 |
| Consensus MFE | -6.86 |
| Energy contribution | -8.17 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
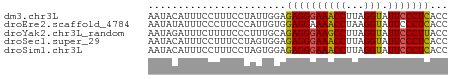
>dm3.chr3L 19379603 75 + 24543557 ---AAUAAAAAUAU---UGCCGGUUG-------UUUGUA---ACCCUUUCAAUUAGUAGCUUGCUUUUGAGUUUGCAAACUUAUAUACAUA ---...........---........(-------((((((---(....(((((..(((.....))).))))).))))))))........... ( -10.50, z-score = -0.02, R) >droSim1.chr3L 18704598 84 + 22553184 UAUAAUAUUAUUGUAGGUGCCAGUUGAUUUUAACUUGAA---ACCCUUUCAAUUGGUAGCUAACUUAUGAGUUAGCAAACUUAUUUA---- ............(((((((((((((((............---......))))))))).(((((((....)))))))..))))))...---- ( -21.77, z-score = -3.44, R) >droSec1.super_29 80820 84 + 700605 UAUAAUAUUAUUGUAGGUGCCGGUUGCUAUUAACUUGAA---ACCCUUUCAAUUGGUAGCUAACUUAUGAGUUAGCAAACUUAUUUA---- ............((((((((((((((.............---.......)))))))).(((((((....)))))))..))))))...---- ( -17.85, z-score = -1.39, R) >droEre2.scaffold_4784 19192051 82 + 25762168 -----AACUACCAUAAAAUAUGGCCGUUUUGGAUUUAAAUUGGUAGUUUCUAACGAUACCAUGAUUCUGAGCUUGCAAUUUUAUUCA---- -----.......(((((((...((.((((.(((((.....(((((...((....))))))).))))).))))..)).)))))))...---- ( -14.40, z-score = -1.00, R) >consensus ___AAUAUUAUUAUAGGUGCCGGUUG_UUUUAACUUGAA___ACCCUUUCAAUUGGUAGCUAACUUAUGAGUUAGCAAACUUAUUUA____ ............((((((((((((((.......................)))))))).(((((((....)))))))..))))))....... ( -6.86 = -8.17 + 1.31)



| Location | 19,379,678 – 19,379,728 |
|---|---|
| Length | 50 |
| Sequences | 5 |
| Columns | 50 |
| Reading direction | forward |
| Mean pairwise identity | 88.40 |
| Shannon entropy | 0.20918 |
| G+C content | 0.42800 |
| Mean single sequence MFE | -14.10 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.52 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.833588 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
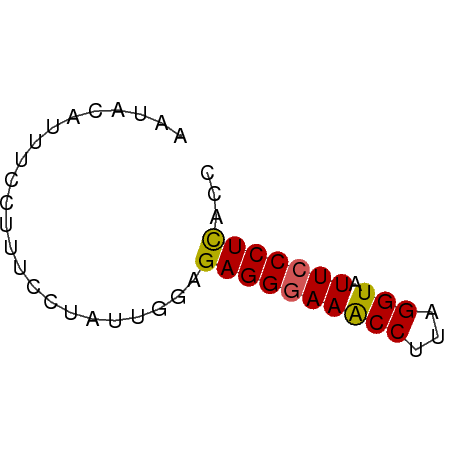
>dm3.chr3L 19379678 50 + 24543557 AAUACAUUUCCUUUCCUAUUGGAGAGGGAAACCUUAGGUAUUCCCUCACC .............(((....)))((((((((((...))).)))))))... ( -15.80, z-score = -2.44, R) >droEre2.scaffold_4784 19192133 50 + 25762168 AAUAUAUUUCCCUUCCCAUUGUGGAGGAAAACCUAAGGUAUUCCCUCACC ....................(((..((((.(((...))).))))..))). ( -11.30, z-score = -1.52, R) >droYak2.chr3L_random 3017530 50 + 4797643 AAUAGAUUUCUUUUCCCUUUGCAGAGGGAAGCCUUAGGUAUUCCCUUACC ...........((((((((....)))))))).....((((......)))) ( -11.80, z-score = -1.01, R) >droSec1.super_29 80904 50 + 700605 AAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACC .............(((....)))((((((((((...))).)))))))... ( -15.80, z-score = -1.99, R) >droSim1.chr3L 18704682 50 + 22553184 AAUACAUUUCCUUUCCUAGUGGAGAGGGAAACCUUAGGUAUUCCCUCACC .............(((....)))((((((((((...))).)))))))... ( -15.80, z-score = -1.99, R) >consensus AAUACAUUUCCUUUCCUAUUGGAGAGGGAAACCUUAGGUAUUCCCUCACC .......................((((((((((...))).)))))))... (-11.64 = -11.52 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:37 2011