| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,348,505 – 19,348,602 |
| Length | 97 |
| Max. P | 0.562622 |

| Location | 19,348,505 – 19,348,602 |
|---|---|
| Length | 97 |
| Sequences | 14 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 85.79 |
| Shannon entropy | 0.31968 |
| G+C content | 0.49686 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -22.01 |
| Energy contribution | -22.57 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.562622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
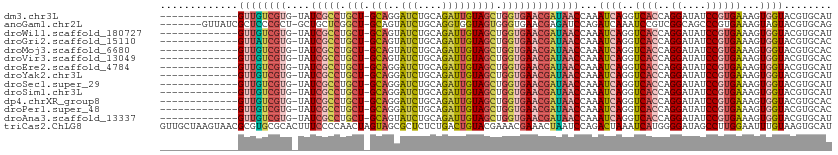
>dm3.chr3L 19348505 97 + 24543557 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.54, R) >anoGam1.chr2L 25235741 103 + 48795086 -------GUUAUCGCUCCCGCU-GCUGCUCGGCU-GCAGUAUCUGCAGGUGGUAGUGGGUGAACGAGAUCCAGAUCAAAUCCGUCGGCAGCCCGUGAAAGUAGUACGUGCAG -------......(((((((((-((..(....((-((((...)))))))..)))))))).((.((.(((.........)))))))))).((.((((.......)))).)).. ( -33.00, z-score = -0.04, R) >droWil1.scaffold_180727 638595 97 + 2741493 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -33.00, z-score = -2.20, R) >droGri2.scaffold_15110 23607603 97 + 24565398 -------------GUUAUCGUG-UAUCGCCUGCU-GCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAC -------------((((((((.-..(((((.(((-(((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -33.00, z-score = -2.36, R) >droMoj3.scaffold_6680 10297262 97 - 24764193 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAC -------------((((((((.-..(((((.(((-(((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -33.00, z-score = -2.10, R) >droVir3.scaffold_13049 12935568 97 + 25233164 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAC -------------((((((((.-..(((((.(((-(((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -33.00, z-score = -2.10, R) >droEre2.scaffold_4784 19160949 97 + 25762168 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.54, R) >droYak2.chr3L 2984480 97 - 24197627 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.54, R) >droSec1.super_29 49994 97 + 700605 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.54, R) >droSim1.chr3L 18673748 97 + 22553184 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.54, R) >dp4.chrXR_group8 7434033 97 + 9212921 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAC -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.48, R) >droPer1.super_48 223890 97 + 581423 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAC -------------((((((((.-..(((((.(((-(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -31.80, z-score = -1.48, R) >droAna3.scaffold_13337 5180829 97 - 23293914 -------------GUUGUCGUG-UAUCGCCUGCU-GCAGUAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU -------------((((((((.-..(((((.(((-(((((........)))))))).)))))))))))))...((((..((((..((....))))))...))))........ ( -33.00, z-score = -2.20, R) >triCas2.ChLG8 3757447 112 - 15773733 GUUGCUAAGUAACGCGUGCGCACUUUCCCCAACUAGUAGCGCUCUCUGACUGUACGAAACGAAACUAAUCCAGACUAAAUCAUGGGGAUAGCCUUGGAAUUUGUAAGUGCAU (((((...)))))(((.(((((((..........))).)))).)....(((.((((((.(((..(((.((((((.....)).))))..)))..)))...))))))))))).. ( -23.80, z-score = 0.49, R) >consensus _____________GUUGUCGUG_UAUCGCCUGCU_GCAGGAUCUGCAGAUUGUAGCUGGUGAACGAUAACCAAAUCAGGUCACCAGGAUAUCCGUGAAAGUGGUACGUGCAU .............((((((((....(((((.(((.(((..(((....))))))))).)))))))))))))...((((..((((..((....))))))...))))........ (-22.01 = -22.57 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:34 2011