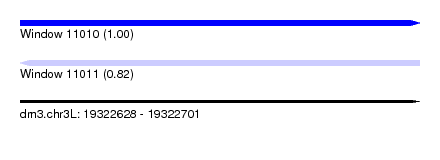
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,322,628 – 19,322,701 |
| Length | 73 |
| Max. P | 0.995274 |

| Location | 19,322,628 – 19,322,701 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 86.47 |
| Shannon entropy | 0.29199 |
| G+C content | 0.49654 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.69 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.995274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
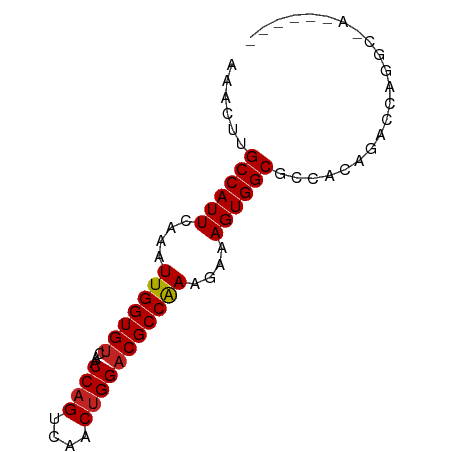
>dm3.chr3L 19322628 73 + 24543557 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACAAAGCAGGCGAAA---- ......((((((....(((((((...((((....)))))))))))....))))))(((........)))....---- ( -25.90, z-score = -3.27, R) >droSim1.chr3L_random 856132 73 + 1049610 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACGGACCAGGCGAAA---- ......((((((....(((((((...((((....)))))))))))....))))))(((........)))....---- ( -25.90, z-score = -2.78, R) >droSec1.super_29 23671 73 + 700605 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACGGACCAGGCGAAA---- ......((((((....(((((((...((((....)))))))))))....))))))(((........)))....---- ( -25.90, z-score = -2.78, R) >droYak2.chr3L 2968012 73 - 24197627 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACAGACCGAGCAACA---- ......((((((....(((((((...((((....)))))))))))....))))))(((.......).))....---- ( -21.80, z-score = -2.15, R) >droEre2.scaffold_4784 19135432 73 + 25762168 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACAGACCAGGCAAUA---- ......((((((....(((((((...((((....)))))))))))....))))))(((........)))....---- ( -25.90, z-score = -3.54, R) >droAna3.scaffold_13337 5158064 69 - 23293914 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACAGCCGAGGC-------- ......((((((....(((((((...((((....)))))))))))....))))))(((........)))-------- ( -24.20, z-score = -2.62, R) >droPer1.super_48 198774 69 + 581423 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACGACAGAAGC-------- ......((((((....(((((((...((((....)))))))))))....))))))..............-------- ( -21.30, z-score = -2.37, R) >dp4.chrXR_group8 7408867 69 + 9212921 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACGACAGAAGC-------- ......((((((....(((((((...((((....)))))))))))....))))))..............-------- ( -21.30, z-score = -2.37, R) >droVir3.scaffold_13049 12326835 77 - 25233164 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCAGCUUCGAAGCAAACGACA ......((((((....(((((((...((((....)))))))))))....))))))....(((....)))........ ( -23.40, z-score = -2.21, R) >droMoj3.scaffold_6680 14501254 59 + 24764193 AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACGGGACGCCGAAGAAAGUGGCGCCA------------------ ......((((((....((((((((..((.......))))))))))....))))))....------------------ ( -19.40, z-score = -1.98, R) >droGri2.scaffold_15110 987484 60 - 24565398 CAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGUCGCCAAAGAAAGUGGCGCCAC----------------- ......((((((....((((((...(((((....)))))))))))....)))))).....----------------- ( -21.30, z-score = -3.46, R) >consensus AAACUUGCCAUUCAAAUUGGUGUCAACCAGUCAACUGGACGCCAAAGAAAGUGGCGCCACAGACCAGGC_A______ ......((((((....(((((((...((((....)))))))))))....))))))...................... (-20.59 = -20.69 + 0.10)

| Location | 19,322,628 – 19,322,701 |
|---|---|
| Length | 73 |
| Sequences | 11 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 86.47 |
| Shannon entropy | 0.29199 |
| G+C content | 0.49654 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -17.85 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.65 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.821418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19322628 73 - 24543557 ----UUUCGCCUGCUUUGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ----....(((........)))((((.((...((((.((((((....))))...)).))))...)).))))...... ( -22.50, z-score = -1.65, R) >droSim1.chr3L_random 856132 73 - 1049610 ----UUUCGCCUGGUCCGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ----....(((........)))((((.((...((((.((((((....))))...)).))))...)).))))...... ( -22.50, z-score = -1.22, R) >droSec1.super_29 23671 73 - 700605 ----UUUCGCCUGGUCCGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ----....(((........)))((((.((...((((.((((((....))))...)).))))...)).))))...... ( -22.50, z-score = -1.22, R) >droYak2.chr3L 2968012 73 + 24197627 ----UGUUGCUCGGUCUGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ----(((((..(((((..((((((((.......))))).)))...)))))..)))))..((((((.....)))))). ( -23.00, z-score = -1.33, R) >droEre2.scaffold_4784 19135432 73 - 25762168 ----UAUUGCCUGGUCUGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ----..(((((.((((..((((((((.......))))).)))...))))(((....))).........))))).... ( -23.90, z-score = -1.91, R) >droAna3.scaffold_13337 5158064 69 + 23293914 --------GCCUCGGCUGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU --------((((((((((.(((((((.......)))))))))))))).((((....))))........)))...... ( -27.60, z-score = -3.06, R) >droPer1.super_48 198774 69 - 581423 --------GCUUCUGUCGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU --------(((........)))((((.((...((((.((((((....))))...)).))))...)).))))...... ( -19.80, z-score = -0.97, R) >dp4.chrXR_group8 7408867 69 - 9212921 --------GCUUCUGUCGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU --------(((........)))((((.((...((((.((((((....))))...)).))))...)).))))...... ( -19.80, z-score = -0.97, R) >droVir3.scaffold_13049 12326835 77 + 25233164 UGUCGUUUGCUUCGAAGCUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ((((((((.......(((((((((((.......))))).))))))...((((....))))....))))))))..... ( -25.90, z-score = -2.07, R) >droMoj3.scaffold_6680 14501254 59 - 24764193 ------------------UGGCGCCACUUUCUUCGGCGUCCCGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ------------------.((((((.........))))))(((((.(.((((....))))...).)))))....... ( -15.10, z-score = -0.35, R) >droGri2.scaffold_15110 987484 60 + 24565398 -----------------GUGGCGCCACUUUCUUUGGCGACCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUG -----------------.....((((.((...(((((((((((....)))))))...))))...)).))))...... ( -20.20, z-score = -2.24, R) >consensus ______U_GCCUGGUCCGUGGCGCCACUUUCUUUGGCGUCCAGUUGACUGGUUGACACCAAUUUGAAUGGCAAGUUU ........(((........)))((((.((...((((.((((((....))))...)).))))...)).))))...... (-17.85 = -18.50 + 0.65)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:31 2011