
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 606,480 – 606,583 |
| Length | 103 |
| Max. P | 0.512852 |

| Location | 606,480 – 606,583 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.78 |
| Shannon entropy | 0.37653 |
| G+C content | 0.40321 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -15.47 |
| Energy contribution | -15.95 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.512852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 606480 103 + 23011544 --AGCAAGAGCAAUACCAACGGCAACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGU-GGUGCUGCAA- --.......(((.(((((..((((.....))))....(((((((((((.......)))))))))))(((..(((.....)))..))).......)-)))).)))..- ( -28.60, z-score = -2.04, R) >droAna3.scaffold_12916 1607910 99 + 16180835 ---GCAACAGAAACACCAACACCACCGGCUGCUAAUGUCUUGUCGGCCAAACCAAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGG-GCUGCUG---- ---.((((((......).........(((((.(((....))).))))).......))))).((((((..(((((................)))))-.))))))---- ( -18.49, z-score = 0.35, R) >droPer1.super_8 3658208 101 - 3966273 --CCCCAGUGCCACAGCAACAGCCACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGCUGCCGCUU---- --.......((..((((((((((....))))......(((((((((((.......)))))))))))(((..(((.....)))..)))....))))))..))..---- ( -26.60, z-score = -2.61, R) >dp4.chr4_group3 2567891 101 - 11692001 --CCCCAGUGCCACAGCAACAGCCACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGCUGCCGCUU---- --.......((..((((((((((....))))......(((((((((((.......)))))))))))(((..(((.....)))..)))....))))))..))..---- ( -26.60, z-score = -2.61, R) >droSim1.chr2L 612862 103 + 22036055 --AGCAAGAGCAAUACCAACGGCAACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGU-GUUGCUGCAA- --.(((...((((((((((((....).))))......(((((((((((.......)))))))))))(((..(((.....)))..)))......))-))))))))..- ( -30.30, z-score = -2.75, R) >droSec1.super_14 589200 103 + 2068291 --AGCAAGAGCAAUACCAACGGCAACAAUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGU-GGUGCUGCAA- --.......(((.(((((..((((.....))))....(((((((((((.......)))))))))))(((..(((.....)))..))).......)-)))).)))..- ( -28.60, z-score = -2.35, R) >droYak2.chr2L 595356 105 + 22324452 AGCGCAACAGCAAUACCAACGGCAACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGU-GGUGCUGCAA- (((((.((((((.((.(((((....).)))).)).)))((((((((((.......)))))))))).(((..(((.....)))..))).....)))-.)))))....- ( -30.10, z-score = -2.19, R) >droEre2.scaffold_4929 662414 103 + 26641161 --AGCAACAGCAAUACCAACGGCAACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGU-GGUGCUGCAA- --.......(((.(((((..((((.....))))....(((((((((((.......)))))))))))(((..(((.....)))..))).......)-)))).)))..- ( -28.60, z-score = -2.11, R) >droMoj3.scaffold_6500 8232027 87 - 32352404 -------------------CAGCAACAGUUGCUAAUGUGUUGUCAACAGAACUCGCGCUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGC-GCUGCGUGUGA -------------------..(((.((((.((.(((.((((((((.(.(....)..).))))))))(((..(((.....)))..)))..))).))-))))..))).. ( -18.40, z-score = 1.24, R) >droVir3.scaffold_12963 19068951 87 + 20206255 -------------------CAGCAACAGUUGCUAAUGUGUUGUCAACAGAACUCGCGUUGACAGCAGCAUCGAAAAUAUUUCAUUUCAAAUUUGC-GCUGCGUGUGA -------------------..(((.((((.((..(((((((((((((.(....)..)))))))))).))).((((........))))......))-))))..))).. ( -22.40, z-score = -0.01, R) >consensus __AGCAAGAGCAAUACCAACAGCAACAGUUGCUAAUGUGUUGUCAACAAAACUCAUGUUGACAGCAGAAUCGAAAAUAUUUCAUUUCAAAUUUGU_GCUGCUGCAA_ ....................((((.(...........((((((((((.........))))))))))(((..(((.....)))..))).........).))))..... (-15.47 = -15.95 + 0.48)

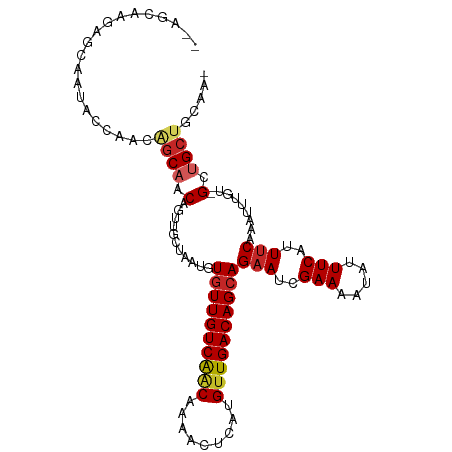
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:18 2011