
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,316,602 – 19,316,715 |
| Length | 113 |
| Max. P | 0.877933 |

| Location | 19,316,602 – 19,316,715 |
|---|---|
| Length | 113 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Shannon entropy | 0.36552 |
| G+C content | 0.47241 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -22.05 |
| Energy contribution | -23.02 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19316602 113 + 24543557 UUUUGGCUGGCAUCUGGCAUUCGCAAUCCAUGAGUACUUCAUUAGGCAAUUUUCGUGGGUCCGGCUCAA------AGGCCCAUUAAAAUUGUCAACUGAGGUCAGUUAAUGAGGCCUUC ....((((..(((.(((((((((.......)))))((((((...(((((((((.(((((.((.......------.))))))).)))))))))...))))))..))))))).))))... ( -36.10, z-score = -2.27, R) >droSim1.chr3L 18632060 113 + 22553184 UUUUGGCUGGCAUCUGGCAUUCGCAAUCCAUGAGCACUUCAUUAGGCAAUUUUCGUGGGUCCGGCUCAA------AGGCCCAUUAAAAUUGUCAACUGAGGUCAGUUAAUGAGGCCUUC ....((((.((.(((((.((.....))))).))))...(((((.(((((((((.(((((.((.......------.))))))).)))))))))(((((....))))))))))))))... ( -37.50, z-score = -2.41, R) >droSec1.super_29 17681 113 + 700605 UUUUGGCUGGCAUCUGGCAUUCGCAAUCCAUGAGCACUUCAUUAGGCAAUUUUCGUGGGUCCGGCUCAA------AGGCACAUUAAAAUUGUCAACUGAGGUCAGUUAAUGAGGCCUUC ....((((.((.(((((.((.....))))).))))...(((((.(((((((((.(((.(.((.......------.))).))).)))))))))(((((....))))))))))))))... ( -30.50, z-score = -0.76, R) >droYak2.chr3L 2961876 106 - 24197627 -------UUUUGGCUGGCAUUCGCAAUCCACGAGCACUUCAUUAGGCAAUUUUCGUGGGUCCGGCUCAA------AAGCCCAUUAAAAUUGUCAACUAAGGUCAGUUAAUGAGGCCUUC -------((((((((((........((((((((((.((.....)))).....))))))))))))).)))------))((((((((..((((((......)).))))))))).))).... ( -29.80, z-score = -1.90, R) >droEre2.scaffold_4784 19129142 106 + 25762168 -------UUUUGGCUGGCACUCGCAAUCCAUGAGCACUUCAUUAGGCAAUUUUCGUGGGUCCGACUCGA------AGGCCCAUUAAAAUUGUCAACUGAGGUCAGUUAAUGAGGCCUUC -------..(((((((((.(((((.........)).........(((((((((.(((((.((.......------.))))))).)))))))))....)))))))))))).......... ( -35.60, z-score = -2.74, R) >droAna3.scaffold_13337 5152283 101 - 23293914 -------UUUUGGCUGGCAUUCGCAAUCCAUGAGCACUUCAUUAGGCAAUUUUCGUGGGUCCGGCUCCA------AGACCCAUUAAAAUUGUCAACUAAGGUCA-----UGAGGCCUCC -------....((((.((....))....(((((.(.(((..((.(((((((((.(((((((.(....).------.))))))).)))))))))))..)))))))-----)).))))... ( -32.70, z-score = -2.55, R) >dp4.chrXR_group8 7402671 101 + 9212921 ------------------UCUGAGGUUCCGUGAGCACUUCAUUAGGCAAUUUUCGUUGGUCCGGCACAGGCACAGAGGCCCACUCAAAUUGUCAACUAAGGUCAGUUAAUGAGGCCUUC ------------------..((((((..(....).))))))...((((((((..((.(((((.((....))...).)))).))..))))))))....((((((.........)))))). ( -28.00, z-score = -0.84, R) >droPer1.super_48 192592 101 + 581423 ------------------UCUGAGGUUCCGUGAGCACUUCAUUAGGCAAUUUUCGUUGGUCCGGCACAGGCACAGAGGCCCACUCAAAUUGUCAACUAAGGUCAGUUAAUGAGGCCUUC ------------------..((((((..(....).))))))...((((((((..((.(((((.((....))...).)))).))..))))))))....((((((.........)))))). ( -28.00, z-score = -0.84, R) >consensus _______UGGCAGCUGGCAUUCGCAAUCCAUGAGCACUUCAUUAGGCAAUUUUCGUGGGUCCGGCUCAA______AGGCCCAUUAAAAUUGUCAACUAAGGUCAGUUAAUGAGGCCUUC ...............................(((..(((((((.(((((((((.(((((((...............))))))).)))))))))((((......))))))))))).))). (-22.05 = -23.02 + 0.97)

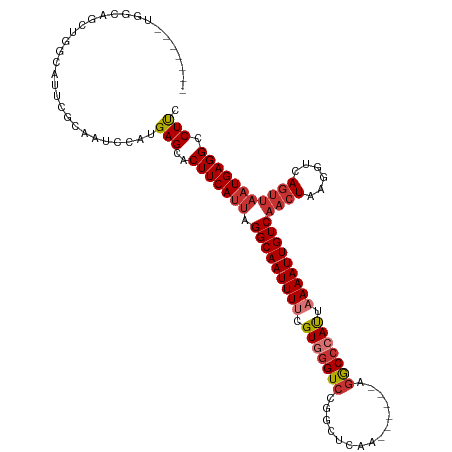
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:29 2011