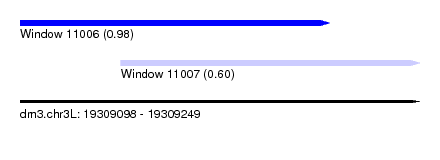
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,309,098 – 19,309,249 |
| Length | 151 |
| Max. P | 0.979026 |

| Location | 19,309,098 – 19,309,215 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Shannon entropy | 0.45853 |
| G+C content | 0.37990 |
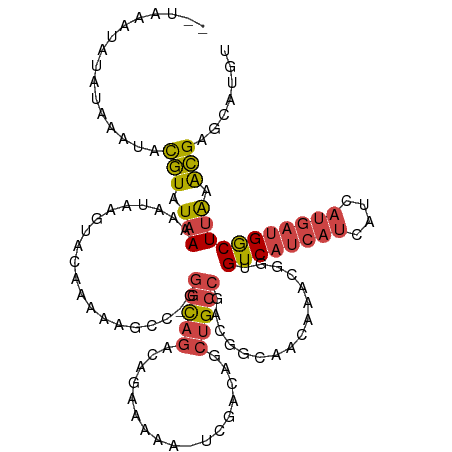
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -12.75 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
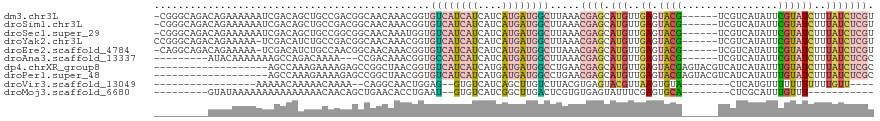
>dm3.chr3L 19309098 117 + 24543557 --AAUAUAUAUAUGUACGUAUAAAAAUAAGUACAAAACGUC-GGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGU --.......((((((.(((.(((..............((((-((.(((...((.....)).....)))))))))............((((((((....))))))))))).))).)))))) ( -30.40, z-score = -2.97, R) >droSim1.chr3L 18624132 119 + 22553184 AAUAAAUAUGUACGUACGUAUAAAAAUAAGUACAAAACGCC-GGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGU .....((((((.(((..((((........)))).....(((-((((((...((.....)).....)))))...(....)...))))((((((((....))))))))....))).)))))) ( -33.00, z-score = -3.37, R) >droSec1.super_29 10237 119 + 700605 AAUAAAUAUGUACGUACGUAUAAAAAUAAGUACAAAACGCC-GGGCAGACAGAAAAAAUCGACAGCUGCCGGCGGCAACAAAUGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGU .....((((((.(((((((((........))))....((((-((.(((...((.....)).....)))))))))..........))((((((((....))))))))....))).)))))) ( -33.70, z-score = -3.16, R) >droYak2.chr3L 2954233 111 - 24197627 --------AAUAAAUACGUAUAAAAAUAAGUAAAAAAAAGCCGGGCAGACAGAAAAA-UCGACAUCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGU --------........(((.(((................((((((((((..((....-))....))))))...(....)...))))((((((((....))))))))))).)))....... ( -27.70, z-score = -3.31, R) >droEre2.scaffold_4784 19121385 110 + 25762168 --------AAUAAAUACGUAUAAAAAUAAGUCCAAAAAGGC-AGGCAGACAGAAAAA-UCGACAUCUGCCAACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGU --------........(((.(((........((........-.((((((..((....-))....))))))...(....)....)).((((((((....))))))))))).)))....... ( -23.40, z-score = -2.14, R) >droMoj3.scaffold_6680 10244404 109 - 24764193 --UAAAUGUAUAUACAUAUAUAAGAAGAAAAAGAUUAAGAGUGUAUAAAAAAAAAAA-AAAACAACAGCUGA--ACACCUGAAUGUGUCAUCGGCUU------GACUCGUGUGAGUAUUU --........(((((((.(.(((...........))).).)))))))..........-........((((((--((((......))))..)))))).------.((((....)))).... ( -15.50, z-score = -0.92, R) >consensus __UAAAUAUAUAAAUACGUAUAAAAAUAAGUACAAAAAGCC_GGGCAGACAGAAAAA_UCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGU ................(((.(((....................(((((.................)))))................((((((((....))))))))))).)))....... (-12.75 = -13.45 + 0.70)

| Location | 19,309,136 – 19,309,249 |
|---|---|
| Length | 113 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.95 |
| Shannon entropy | 0.68479 |
| G+C content | 0.43037 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -8.47 |
| Energy contribution | -9.16 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19309136 113 + 24543557 -CGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG------UCGUCAUAUUCGUAUCUUUAUCUCGU -((((..(((.((.....(((((((((((((..(....)...))))((((((((....)))))))).......))).)))))).....------)))))......(((....))))))). ( -29.70, z-score = -0.85, R) >droSim1.chr3L 18624172 113 + 22553184 -CGGGCAGACAGAAAAAAUCGACAGCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG------UCGUCAUAUUCGUAUCUUUAUCUCGU -((((..(((.((.....(((((((((((((..(....)...))))((((((((....)))))))).......))).)))))).....------)))))......(((....))))))). ( -29.70, z-score = -0.85, R) >droSec1.super_29 10277 113 + 700605 -CGGGCAGACAGAAAAAAUCGACAGCUGCCGGCGGCAACAAAUGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG------UCGUCAUAUUCGUAUCUUUAUCUCGU -((((..(((.((.....(((((((((((((..(....)...))))((((((((....)))))))).......))).)))))).....------)))))......(((....))))))). ( -29.10, z-score = -0.57, R) >droYak2.chr3L 2954265 113 - 24197627 CCGGGCAGACAGAAAAA-UCGACAUCUGCCGACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG------UCGUCAUAUUCGUAUCUUUAUCUCGU (((((((((..((....-))....))))))...(....)...))).((((((((....))))))))....(((((.(((..((.((((------..........))))))..)))))))) ( -32.90, z-score = -1.89, R) >droEre2.scaffold_4784 19121417 112 + 25762168 -CAGGCAGACAGAAAAA-UCGACAUCUGCCAACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG------UCGUCAUAUUCGUAUCUUUAUCUCGU -..((((((..((....-))....))))))...(....).......((((((((....))))))))....(((((.(((..((.((((------..........))))))..)))))))) ( -29.60, z-score = -1.81, R) >droAna3.scaffold_13337 5145308 102 - 23293914 ---------AUACAAAAAAAGCCAGACAAAA---CCGACAAACGGUGCCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG------UCGUCAUAUUCGUAUCUUUAUCUCGC ---------...........((.(((.((((---(((.....))))((((((((....))))))))....(((((.(((.(((.....------))).))).)))))...))).))).)) ( -24.50, z-score = -2.26, R) >dp4.chrXR_group8 7395772 101 + 9212921 -------------------AGCCAAAGAAAAGAGCCGGCUAACGGUGUCAUCAUCAUGAUGAUGGCCUGAACGAGCAUGUUGAGUACGAGUACGUCAUCAUAUUUGUAUCUUUAUCUCGC -------------------((((.............))))..(((.((((((((....)))))))))))..((((.(((..((.((((((((.(....).))))))))))..))))))). ( -29.12, z-score = -1.69, R) >droPer1.super_48 185721 101 + 581423 -------------------AGCCAAAGAAAAGAGCCGGCUAACGGUGUCAUCAUCAUGAUGAUGGCCUGAACGAGCAUGUUGAGUACGAGUACGUCAUCAUAUUUGUAUCUUUAUCUCGC -------------------((((.............))))..(((.((((((((....)))))))))))..((((.(((..((.((((((((.(....).))))))))))..))))))). ( -29.12, z-score = -1.69, R) >droVir3.scaffold_13049 12892312 87 + 25233164 -----------------AAAAACAAAAACAAAA--CAGGCAACUGGAG--GUGUCAUCAGCUUGUCUUACGUGAGUACGUUAAGUGUA--------CUCAUGUUUUUUUUUUUGUU---- -----------------...((((((((.((((--(((....)))(((--.((....)).))).....(((((((((((.....))))--------))))))))))).))))))))---- ( -26.40, z-score = -3.55, R) >droMoj3.scaffold_6680 10244444 90 - 24764193 ---------GUAUAAAAAAAAAAAAAAACAACAGCUGAACACCUGAAU--GUGUCAUCGGCUUGACUCGUGUGAGUAUUUCGAGUGCA--------CUCGCAUUUGUUU----------- ---------................((((((.((((((((((......--))))..))))))......((((((((...........)--------)))))))))))))----------- ( -19.50, z-score = -1.11, R) >consensus _________CAGAAAAAAUAGACAAAUACAAACGGCAACAAACGGUGUCAUCAUCAUCAUGAUGGCUUAAACGAGCAUGUUGAGUACG______UCGUCAUAUUCGUAUCUUUAUCUCGU ..............................................((((((((....)))))))).....((((.(((..((.((((................))))))..))))))). ( -8.47 = -9.16 + 0.69)
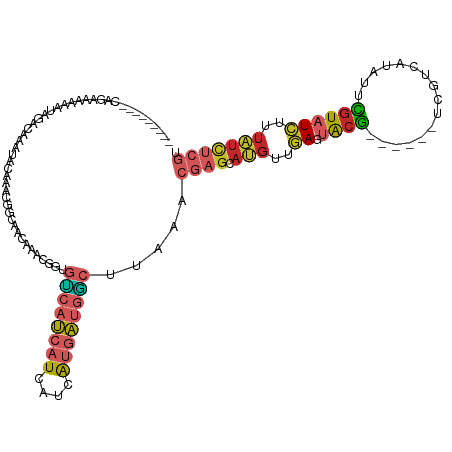
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:27 2011