| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,279,658 – 19,279,750 |
| Length | 92 |
| Max. P | 0.895307 |

| Location | 19,279,658 – 19,279,750 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.09 |
| Shannon entropy | 0.60500 |
| G+C content | 0.35250 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -9.48 |
| Energy contribution | -10.36 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
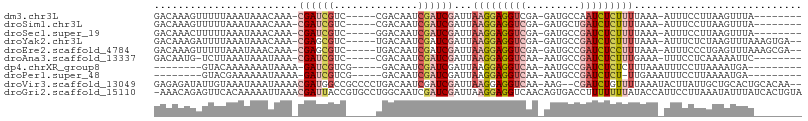
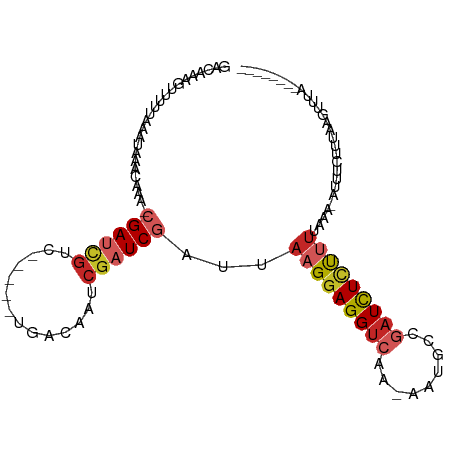
>dm3.chr3L 19279658 92 + 24543557 GACAAAGUUUUUAAAUAAACAAA-CGAUCGUC-----CGACAAUCGAUCGAUUAAGGAGGUCGA-GAUGCCAAUCUCUUUUAAA-AUUUCCUUAAGUUUA-------- ...............(((((...-((((((..-----.......)))))).((((((((((.((-(((....))))).......-)))))))))))))))-------- ( -21.60, z-score = -2.49, R) >droSim1.chr3L 18589127 92 + 22553184 GACAAAGUUUUUAAAUAAACAAA-CGAUCGUC-----CGACAAUCGAUCGAUUAAGGAGGUCGA-GAUGCUGAUCUCUUUUAAA-AUUUCCUUAAGUUUA-------- ...............(((((...-((((((..-----.......)))))).((((((((((.((-(((....))))).......-)))))))))))))))-------- ( -20.90, z-score = -1.67, R) >droSec1.super_19 1233463 92 + 1257711 GACAAACUUUUUAAAUAAACAAA-CGAUCGUC-----GGACAAUCGAUCGAUUAAGGAGGUCGA-GAUGCCGAUCUCUUUUAAA-AUUUCCUUAAGUUUA-------- ...((((((...((((.......-((((((..-----.......))))))...((((((((((.-.....))))))))))....-))))....)))))).-------- ( -22.00, z-score = -1.92, R) >droYak2.chr3L 2918070 98 - 24197627 GACAAAGAUUUUAAAUAAACAAA-CGAGCGUC-----UGACAAUCGAUCGAUUAAGGAGGUCGA-GAUGCCGAUCUCUUUUAAA-AUUUCUCUAAGUUUAAAGUGA-- .......(((((((((.......-(((.((..-----.......)).)))...((((((((((.-.....))))))))))....-..........)))))))))..-- ( -19.30, z-score = -0.32, R) >droEre2.scaffold_4784 19091538 98 + 25762168 GACAAAGUUUUUAAAUAAACAAA-CGAGCGUC-----UGACAAUCGAUCGAUUAAGGAGGUCGA-GAUGCCGAUCUCCUUUAAA-AUUUCCCUGAGUUUAAAGCGA-- ......((((......))))...-...((...-----.(((((((....))))((((((((((.-.....))))))))))....-..........)))....))..-- ( -21.90, z-score = -1.10, R) >droAna3.scaffold_13337 5114474 91 - 23293914 GACAAUG-UCUUAAAUAAAUAAA-CGAUCGUC-----CGACAAUCGAUCGAUUAAGGAGGUCAA-AAUGCCGAUCUCUUUGAAA-UUUCCUCAAAAAUUC-------- (((...)-)).............-((((((..-----.......)))))).(((((((((((..-......)))))))))))..-...............-------- ( -19.70, z-score = -2.08, R) >dp4.chrXR_group8 7367161 84 + 9212921 --------GUACAAAAAAAUAAAA-GAUCGUCG-----GACAAUCGAUCGAUUAAGGAGGUCAA-AAUGCCGAUCUCUCUUUAAAUUUCCUUAAAAUGA--------- --------................-((((((((-----......))).)))))(((((((((..-......)))))).)))..................--------- ( -14.70, z-score = -0.87, R) >droPer1.super_48 157281 83 + 581423 --------GUACGAAAAAAUAAAA-GAUCGUCG-----GACAAUCGAUCGAUUAAGGAGGUCAA-AAUGCCGAUCUCU-UUGAAAUUUCCUUAAAAUGA--------- --------....((((........-(((((...-----......)))))..(((((((((((..-......)))))))-))))..))))..........--------- ( -18.80, z-score = -1.57, R) >droVir3.scaffold_13049 12858606 103 + 25233164 GAGAGAUAUUGUAAAUAAAUAAAACGAUGGCCGCCCCUGACAAUCGAUCGAUUAAGGAGGUCAA-AAG--CGAUCUGUUUUAAAUACUUAUUGCUGCACUGCACAA-- ........((((.......(((((((((.((.((((((...((((....)))).))).)))...-..)--).))).))))))..........((......))))))-- ( -19.70, z-score = -0.57, R) >droGri2.scaffold_15110 23511186 107 + 24565398 -AAACAGAGUUCACAAAAAUUAAACGAUUACCGUGCCUGGCAAUCGAUCGAUUAAGGAGGUCAACAGUGACCUUUUUUUAUACCAUUCCUUAAAUAUUUAUCACUGUA -..((((.................(((((.(((....))).))))).....((((((((((((....)))))..............)))))))..........)))). ( -21.03, z-score = -1.77, R) >consensus GACAAAGUUUUUAAAUAAACAAA_CGAUCGUC_____UGACAAUCGAUCGAUUAAGGAGGUCAA_AAUGCCGAUCUCUUUUAAA_AUUUCUUUAAGUUUA________ ........................((((.(((......))).)))).......(((((((((.........)))))))))............................ ( -9.48 = -10.36 + 0.88)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:24 2011