| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,213,447 – 19,213,569 |
| Length | 122 |
| Max. P | 0.977820 |

| Location | 19,213,447 – 19,213,544 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.54 |
| Shannon entropy | 0.46773 |
| G+C content | 0.39432 |
| Mean single sequence MFE | -14.33 |
| Consensus MFE | -8.36 |
| Energy contribution | -8.44 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
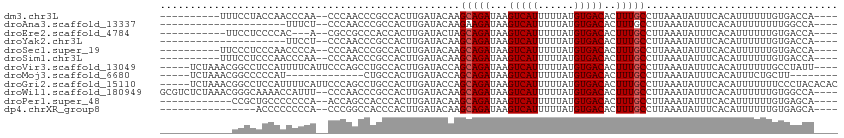
>dm3.chr3L 19213447 97 + 24543557 ----------UUUCCUACCAACCCAA--CCCAACCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCA---- ----------................--......................(((((...(((((......)))))..)))))..........(((((.....)))))...---- ( -13.40, z-score = -2.75, R) >droAna3.scaffold_13337 5052145 86 - 23293914 ---------------------UUUCU--CCCAACCCGCCACUUGAUACAAGAAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUUUGGCCA---- ---------------------.....--........((((((((...)))).......(((((......))))).............................))))..---- ( -11.90, z-score = -1.56, R) >droEre2.scaffold_4784 19024131 93 + 25762168 -----------UUCCUCCCCAC---A--CGCCGCCCACCACUUGAUACUAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCA---- -----------...........---.--......................(((((...(((((......)))))..)))))..........(((((.....)))))...---- ( -13.40, z-score = -2.50, R) >droYak2.chr3L 2845351 86 - 24197627 ---------------------UUCCU--CCCAACCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCA---- ---------------------.....--......................(((((...(((((......)))))..)))))..........(((((.....)))))...---- ( -13.40, z-score = -2.62, R) >droSec1.super_19 1167505 97 + 1257711 ----------UUCCCUCCCAACCCCA--CCCAACCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCA---- ----------................--......................(((((...(((((......)))))..)))))..........(((((.....)))))...---- ( -13.40, z-score = -2.94, R) >droSim1.chr3L 18520327 97 + 22553184 ----------UUUCCUCCCAACCCAA--CCCAACCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCA---- ----------................--......................(((((...(((((......)))))..)))))..........(((((.....)))))...---- ( -13.40, z-score = -2.73, R) >droVir3.scaffold_13049 13796887 104 + 25233164 -----UCUAAACGGCCUCCAUUUUCAUUCCCAGCCUGCCACUUGAUACCAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGCCUAUU---- -----.......(((...................((((............))))....(((((......))))).....)))...........................---- ( -13.10, z-score = -0.75, R) >droMoj3.scaffold_6680 11222617 87 - 24764193 -----UCUAAACGGCCCCCAU-------------CUGCCACUUGAUACCAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUCUGCUU-------- -----.......(((....((-------------((((............))))))..(((((......))))).....))).......................-------- ( -15.70, z-score = -2.34, R) >droGri2.scaffold_15110 9122953 108 - 24565398 -----UCUAAACGGCCUCCAUUUUCAUUCCCAGCCUGCCACUUGAUACCAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUUCCCUACACAC -----.......(((...................((((............))))....(((((......))))).....)))............................... ( -13.10, z-score = -1.49, R) >droWil1.scaffold_180949 3352728 107 - 6375548 GCGUCUCUAAACGGGCAAAACCAUUU--CCCAACCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGGCCA---- ............(((.(((....)))--))).....((((((((...)))(((((...(((((......)))))..))))).....................)))))..---- ( -19.00, z-score = -0.76, R) >droPer1.super_48 92468 95 + 581423 ------------CCGCUGCCCCCCCA--ACCAGCCACCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGAGCA---- ------------..((((........--..))))................(((((...(((((......)))))..))))).........((((((.....))))))..---- ( -16.90, z-score = -2.62, R) >dp4.chrXR_group8 7302905 91 + 9212921 ----------------ACCCCCCCCA--CCCGGCCACCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGAGCA---- ----------------..........--...(((......((((...)))).......(((((......))))).....)))........((((((.....))))))..---- ( -15.20, z-score = -2.52, R) >consensus ____________GCCCCCCAACCCCA__CCCAACCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCA____ ..................................................(((((...(((((......)))))..)))))................................ ( -8.36 = -8.44 + 0.08)

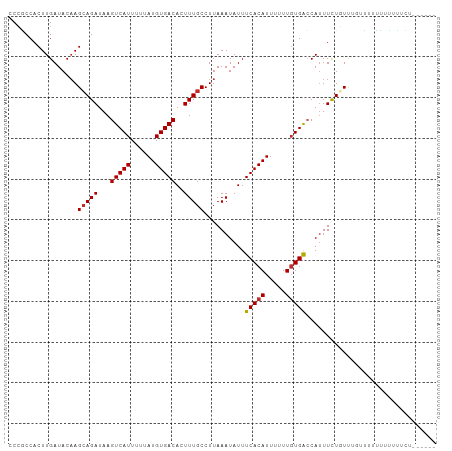
| Location | 19,213,468 – 19,213,569 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.14 |
| Shannon entropy | 0.33198 |
| G+C content | 0.33424 |
| Mean single sequence MFE | -18.10 |
| Consensus MFE | -12.13 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.973961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19213468 101 + 24543557 CCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCAUUUCUGUUGUUUUUUUUUCUUCUUU---- ................((((((...(((((....((((((....((......))....)))))).....)))))....)))))).................---- ( -16.50, z-score = -2.57, R) >droAna3.scaffold_13337 5052155 84 - 23293914 CCCGCCACUUGAUACAAGAAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUUUGGCCAUUUUUUUC--------------------- ...((((((((...)))).......(((((......))))).............................))))..........--------------------- ( -11.90, z-score = -1.61, R) >droEre2.scaffold_4784 19024148 96 + 25762168 CCCACCACUUGAUACUAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCAUUUCUGUUUUUUUCUUCUUU--------- ..........((....((((((...(((((....((((((....((......))....)))))).....)))))....))))))....))......--------- ( -17.30, z-score = -3.78, R) >droYak2.chr3L 2845361 100 - 24197627 CCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCACUUCUGUU-UUUUUUUUCCUUCUUU---- ...............(((((((...(((((....((((((....((......))....)))))).....)))))....))))))-)...............---- ( -16.80, z-score = -3.43, R) >droSec1.super_19 1167526 99 + 1257711 CCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCAUUUCUGUUGGUUUUUUCUUCUUU------ ...((((((((...))))((((...(((((....((((((....((......))....)))))).....)))))....)))).))))............------ ( -17.40, z-score = -2.11, R) >droSim1.chr3L 18520348 101 + 22553184 CCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCAUUUCUGUUGGUUUUUUUUCUUCUUU---- ...((((((((...))))((((...(((((....((((((....((......))....)))))).....)))))....)))).))))..............---- ( -17.40, z-score = -2.19, R) >droWil1.scaffold_180949 3352759 105 - 6375548 CCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGGCCAUUUUUUUUUUUUCUCUUCUCUUUGCCUCU ...((((((((...)))(((((...(((((......)))))..))))).....................)))))............................... ( -15.30, z-score = -1.78, R) >droPer1.super_48 92487 98 + 581423 CCACCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGAGCAUUUUUUUCUGAGAUGGGGUGUG------- .(((((...........(((((...(((((......)))))..))))).........((((((.....))))))((((((.....)))))))))))..------- ( -25.60, z-score = -2.84, R) >dp4.chrXR_group8 7302920 98 + 9212921 CCACCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGAGCAUUUUUUUCUGGGAUGGGGUGUG------- .(((((...........(((((...(((((......)))))..))))).........((((((.....))))))((((((.....)))))))))))..------- ( -24.70, z-score = -2.03, R) >consensus CCCGCCACUUGAUACAAGCAGAUAAGUCAUUUUUAUGUGACACUUUGCCUUAAAUAUUUCACAUUUUUUGUGACCAUUUCUGUUUGUUUUUUUUUUUCU______ .................(((((...(((((......)))))..)))))..........(((((.....)))))................................ (-12.13 = -12.18 + 0.05)

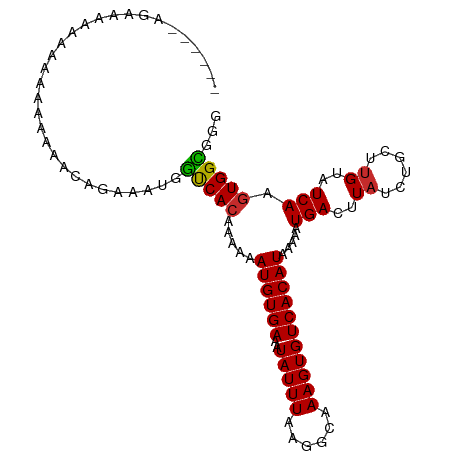
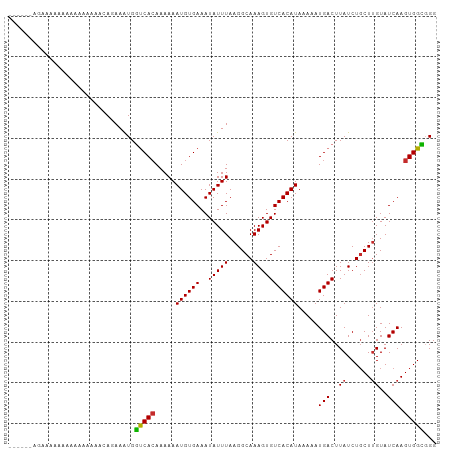
| Location | 19,213,468 – 19,213,569 |
|---|---|
| Length | 101 |
| Sequences | 9 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.14 |
| Shannon entropy | 0.33198 |
| G+C content | 0.33424 |
| Mean single sequence MFE | -17.07 |
| Consensus MFE | -12.69 |
| Energy contribution | -12.32 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19213468 101 - 24543557 ----AAAGAAGAAAAAAAAACAACAGAAAUGGUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGCGGG ----...............((((((((...(((((......((((((..(((((......))))))))))).....)))))..)))).))))............. ( -17.10, z-score = -1.65, R) >droAna3.scaffold_13337 5052155 84 + 23293914 ---------------------GAAAAAAAUGGCCAAAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUUCUUGUAUCAAGUGGCGGG ---------------------..........((((......((((((..(((((......))))))))))).....(((..((......))..)))..))))... ( -12.40, z-score = -0.68, R) >droEre2.scaffold_4784 19024148 96 - 25762168 ---------AAAGAAGAAAAAAACAGAAAUGGUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUAGUAUCAAGUGGUGGG ---------..............((((...(((((......((((((..(((((......))))))))))).....)))))..))))(((.(((...))).))). ( -15.10, z-score = -1.27, R) >droYak2.chr3L 2845361 100 + 24197627 ----AAAGAAGGAAAAAAAA-AACAGAAGUGGUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGCGGG ----................-..((((...(((((......((((((..(((((......))))))))))).....)))))..))))(((((........))))) ( -15.60, z-score = -0.99, R) >droSec1.super_19 1167526 99 - 1257711 ------AAAGAAGAAAAAACCAACAGAAAUGGUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGCGGG ------.............((..((....))(((((...((((((...)))))).(((((.(..((((........))))...).))))).......))))))). ( -17.80, z-score = -1.37, R) >droSim1.chr3L 18520348 101 - 22553184 ----AAAGAAGAAAAAAAACCAACAGAAAUGGUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGCGGG ----...............((..((....))(((((...((((((...)))))).(((((.(..((((........))))...).))))).......))))))). ( -17.80, z-score = -1.50, R) >droWil1.scaffold_180949 3352759 105 + 6375548 AGAGGCAAAGAGAAGAGAAAAAAAAAAAAUGGCCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGCGGG ...............................(((((...((((((...)))))).(((((.(..((((........))))...).))))).......)))))... ( -16.60, z-score = -0.59, R) >droPer1.super_48 92487 98 - 581423 -------CACACCCCAUCUCAGAAAAAAAUGCUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGGUGG -------..((((((...........(((((.(((((.....)))))..))))).(((((.(..((((........))))...).))))).......).))))). ( -20.60, z-score = -1.79, R) >dp4.chrXR_group8 7302920 98 - 9212921 -------CACACCCCAUCCCAGAAAAAAAUGCUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGGUGG -------..((((((...........(((((.(((((.....)))))..))))).(((((.(..((((........))))...).))))).......).))))). ( -20.60, z-score = -1.61, R) >consensus ______AGAAAAAAAAAAAAAAACAGAAAUGGUCACAAAAAAUGUGAAAUAUUUAAGGCAAAGUGUCACAUAAAAAUGACUUAUCUGCUUGUAUCAAGUGGCGGG ...............................(((((.....((((((..(((((......))))))))))).....(((..((......))..))).)))))... (-12.69 = -12.32 + -0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:20 2011