| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,181,772 – 19,181,867 |
| Length | 95 |
| Max. P | 0.633807 |

| Location | 19,181,772 – 19,181,867 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Shannon entropy | 0.21477 |
| G+C content | 0.44098 |
| Mean single sequence MFE | -22.81 |
| Consensus MFE | -18.67 |
| Energy contribution | -18.81 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.633807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19181772 95 + 24543557 AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAACAACAAC------------ ...............((((((((((((...............(((((.....)))))....((...))...)))))))))...))).........------------ ( -21.40, z-score = -1.67, R) >droSim1.chr3L 18488768 95 + 22553184 AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAACAACAAC------------ ...............((((((((((((...............(((((.....)))))....((...))...)))))))))...))).........------------ ( -21.40, z-score = -1.67, R) >droSec1.super_19 1137316 95 + 1257711 AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAACAACAAC------------ ...............((((((((((((...............(((((.....)))))....((...))...)))))))))...))).........------------ ( -21.40, z-score = -1.67, R) >droEre2.scaffold_4784 18993838 94 + 25762168 AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAA-GCAAUGCACAUAGGCACGUCCUAGCACCAACAAC------------ ...............((((((((((((...............(((((.....)))))...-((...))...)))))))))...))).........------------ ( -21.40, z-score = -1.52, R) >droAna3.scaffold_13337 5022276 101 - 23293914 AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUGGCAACAACACCACCGCC------ ...............((((((((((((...............(((((.....)))))....((...))...)))))))))...)))...............------ ( -21.40, z-score = -0.23, R) >droPer1.super_10659 502 107 + 1024 AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUACGUCCUGCGUCCGACGUCCUGCC ....(((........)))..((((((((((.....)))....(((((.....))))).....))).)))).((((.(((((..(((.....)))..))))))))).. ( -27.20, z-score = -0.92, R) >droGri2.scaffold_15110 9091007 94 - 24565398 AAACAGCCGAC-GAGGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAA-GCAGCGAGCAUAGGCACGUUCUAACAACAACAAAC----------- ....((((...-..))))(((((((((...............(((((.....)))))...-((.....)).)))))))))................----------- ( -25.50, z-score = -2.89, R) >consensus AAACAGCCGACAGAAGCUACGUGCUUGGAAAAUAAUUUAAAAGCUCAAAAGGUGAGCAAAAGCAAUGCACAUAGGCACGUCCUAGCAACAACAAC____________ ....(((........)))(((((((((...............(((((.....)))))....((...))...)))))))))........................... (-18.67 = -18.81 + 0.14)

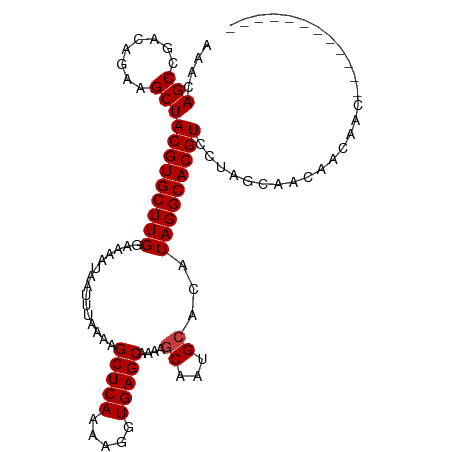
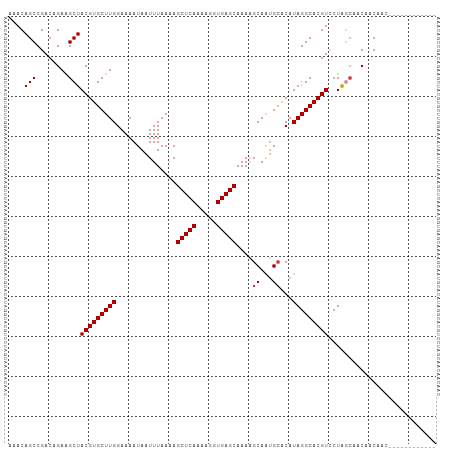
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:15 2011