| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,128,299 – 19,128,419 |
| Length | 120 |
| Max. P | 0.565921 |

| Location | 19,128,299 – 19,128,419 |
|---|---|
| Length | 120 |
| Sequences | 9 |
| Columns | 127 |
| Reading direction | forward |
| Mean pairwise identity | 65.30 |
| Shannon entropy | 0.68586 |
| G+C content | 0.39231 |
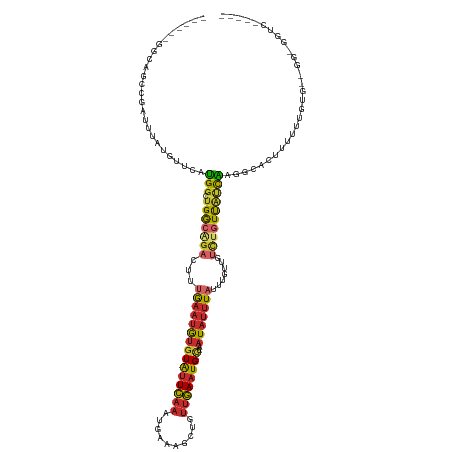
| Mean single sequence MFE | -28.56 |
| Consensus MFE | -9.62 |
| Energy contribution | -9.49 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
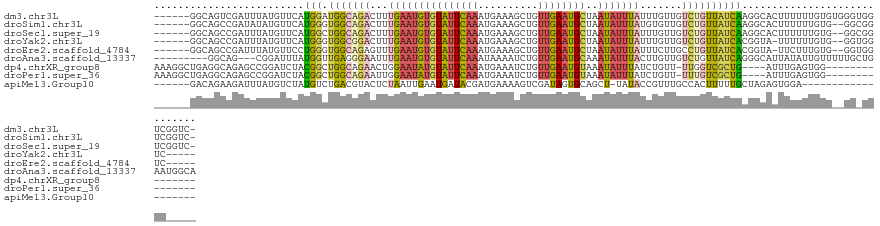
>dm3.chr3L 19128299 120 + 24543557 ------GGCAGUCGAUUUAUGUUCAUGGAUGGCAGACUUUGAAUGUGUAUUCAAAUGAAAGCUGUUGAAUGCUAAUAUUUAUUUGUUGUCUGUUAUCAAGGCACUUUUUUGUGUGGGUGGUCGGUC- ------.....((((((...(..(...((((((((((..(((((((((((((((.(....)...))))))))..)))))))......))))))))))...((((......)))))..)))))))..- ( -31.40, z-score = -1.89, R) >droSim1.chr3L 18434847 118 + 22553184 ------GGCAGCCGAUAUAUGUUCAUGGGUGGCAGACUUUGAAUGUGUAUUCAAAUGAAAGCUGUUGAAUGCUAAUAUUUAUGUGUUGUCUGUUAUCAAGGCACUUUUUUGUG--GGCGGUCGGUC- ------(((.(((.(((...((.(...((((((((((..(((((((((((((((.(....)...))))))))..)))))))......))))))))))...).)).....))).--))).)))....- ( -34.70, z-score = -2.15, R) >droSec1.super_19 1084307 118 + 1257711 ------GGCAGCCGAUUUAUGUUCAUGGCUGGCAGACUUUGAAUGUGUAUUCAAAUGAAAGCUGUUGAAUGCUAAUAUUUAUUUGUUGUCUGUUAUCAAGGCACUUUUUUGUG--GGCGGUCGGUC- ------(((.(((............(((.((((((((..(((((((((((((((.(....)...))))))))..)))))))......)))))))))))...(((......)))--))).)))....- ( -31.00, z-score = -1.04, R) >droYak2.chr3L 2737651 113 - 24197627 ------GGCAGCCGAUUUAUGUUCAUGGGUGGCGGACUUUGAAUGUGUAUUCAAAUGAAAGCUGUUGAAUGCUAAUAUUUAUUUGUUGUCUGUUAUCACGGUA-UUUUUUGUG--GGUGGUC----- ------(((.(((.((.........((((((((((((..(((((((((((((((.(....)...))))))))..)))))))......)))))))))).))...-......)).--))).)))----- ( -29.17, z-score = -1.79, R) >droEre2.scaffold_4784 18940575 113 + 25762168 ------GGCAGCCGAUUUAUGUUCCUGGGUGGCAGAGUUUGAAUGUGUAUUCAAAUGAAAGCUGUUGAAUGCUAAUAUUUAUUUCUUGCCUGUUAUCACGGUA-UUCUUUGUG--GGUGGUC----- ------(((.(((.((........(((((((((((.((.(((((((((((((((.(....)...))))))))..)))))))......)))))))))).)))..-......)).--))).)))----- ( -27.99, z-score = -0.77, R) >droAna3.scaffold_13337 4969451 115 - 23293914 ---------GGCAG---CGGAUUUAUGGUUGAGGGAAUUUGAAUGUGUAUUCAAAUAAAAUCUGUUGAAUGCAAAUAUUUACUUGUUGUCUGUUAUCAGGGCAUUAUAUUGUUUUUGCUGAAUGGCA ---------..(((---(((((((....(((((...((......))...)))))...))))))))))(((((.((((......)))).((((....)))))))))..........((((....)))) ( -23.10, z-score = 0.09, R) >dp4.chrXR_group8 7196601 107 + 9212921 AAAGGCUGAGGCAGAGCCGGAUCUACGGCUGGCAGAACUGGAAUAUGUAUUCAAAUGAAAUCUGUUGAAUGUAAAUAUUUAUCUGUU-UUGGUCGCUG----AUUUGAGUGG--------------- ...(((..((((((((((((..(....)))))).......((((((.(((((((..(....)..)))))))...)))))).))))))-)..)))(((.----.....)))..--------------- ( -30.40, z-score = -2.37, R) >droPer1.super_36 708801 107 - 818889 AAAGGCUGAGGCAGAGCCGGAUCUACGGCUGGCAGAAUUGGAAUAUGUAUUCAAAUGAAAUCUGUUGAAUGUAAAUAUUUAUCUGUU-UUUGUCGCUG----AUUUGAGUGG--------------- ....(((.(((((((((((......)))))(((((((...((((((.(((((((..(....)..)))))))...))))))......)-)))))).)))----.))).)))..--------------- ( -28.00, z-score = -1.73, R) >apiMel3.Group10 9279310 101 + 11440700 ------GACAGAAGAUUUAUGUCUAUGUCUGACGUACUCUAAUUGAAUGAUACGAUGAAAAGUCGAUAGUGCAGCU-UAUACCGUUUGCCACUUUUUGCUAGAGUGGA------------------- ------((((..((((....)))).))))....((((((.....))......((((.....))))...))))....-...........(((((((.....))))))).------------------- ( -21.30, z-score = -0.41, R) >consensus ______GGCAGCCGAUUUAUGUUCAUGGCUGGCAGACUUUGAAUGUGUAUUCAAAUGAAAGCUGUUGAAUGCUAAUAUUUAUUUGUUGUCUGUUAUCAAGGCACUUUUUUGUG__GG_GGUC_____ .........................(((.(((((((...(((((((((((((((..........))))))))..))))))).......))))))))))............................. ( -9.62 = -9.49 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:11 2011