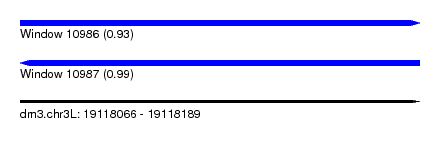
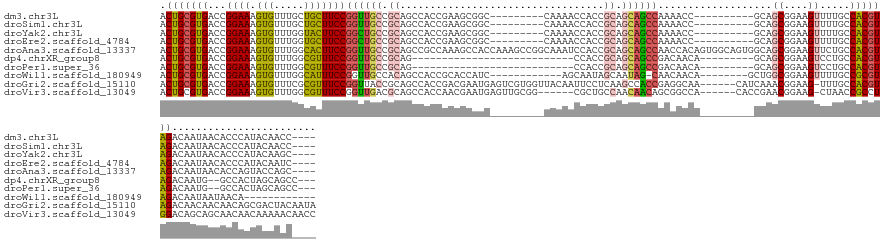
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,118,066 – 19,118,189 |
| Length | 123 |
| Max. P | 0.987545 |

| Location | 19,118,066 – 19,118,189 |
|---|---|
| Length | 123 |
| Sequences | 10 |
| Columns | 146 |
| Reading direction | forward |
| Mean pairwise identity | 69.01 |
| Shannon entropy | 0.61562 |
| G+C content | 0.56927 |
| Mean single sequence MFE | -49.59 |
| Consensus MFE | -22.57 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
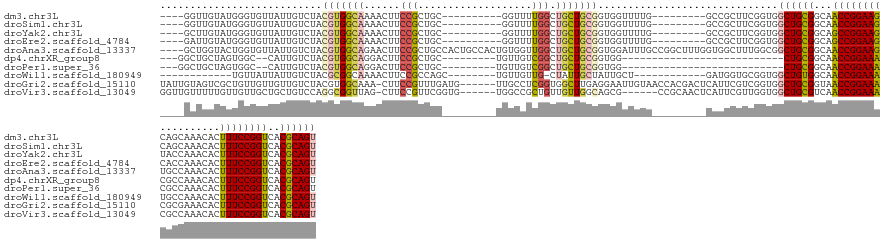
>dm3.chr3L 19118066 123 + 24543557 ----GGUUGUAUGGGUGUUAUUGUCUACGUGGCAAAACUUCCGCUGC----------GGUUUUGGCUGCUGCGGUGGUUUUG---------GCCGCUUCGGUGGCUGCGGCAACCGGAAGCAGCAAACACUUUCCGGUCACGCAGU ----.((..(.(((..(....)..))).)..))((((((.((((.((----------(((....))))).)))).))))))(---------(((((....))))))(((...((((((((..........))))))))..)))... ( -52.00, z-score = -2.08, R) >droSim1.chr3L 18424409 123 + 22553184 ----GGUUGUAUGGGUGUUAUUGUCUACGUGGCAAAACUUCCGCUGC----------GGUUUUGGCUGCUGCGGUGGUUUUG---------GCCGCUUCGGUGGCUGCGGCAACCGGAAGCAGCAAACACUUUCCGGUCACGCAGU ----.((..(.(((..(....)..))).)..))((((((.((((.((----------(((....))))).)))).))))))(---------(((((....))))))(((...((((((((..........))))))))..)))... ( -52.00, z-score = -2.08, R) >droYak2.chr3L 2726811 123 - 24197627 ----GCUUGUAUGGGUGUUAUUGUCUACGUGGCAAAACUUCCGCUGC----------GGUUUUGGCUGCUGCGGUGGUUUUG---------GCCGCUUCGGUGGCUGCGGCAGCCGGAAGUACCAAACACUUUCCGGUCACGCAGU ----((.((..((((((((.(((((.....))))).(((((((((((----------.((...(((..(((.((((((....---------)))))).)))..))))).)))).)))))))....)))))...)))..)).))... ( -53.00, z-score = -2.37, R) >droEre2.scaffold_4784 18929875 123 + 25762168 ----GAUUGUAUGGGUGUUAUUGUCUACGUGGCAAAACUUCCGCUGC----------GGUUUUGGCUGCUGCGGUGGUUUUG---------GCCGCUUCGGUGGCUGCGGCAGCCGGAAGCACCAAACACUUUCCGGUCACGCAGU ----(((((...(((((((.(((((.....)))))..((((((((((----------.((...(((..(((.((((((....---------)))))).)))..))))).)))).)))))).....)))))))..)))))....... ( -53.00, z-score = -2.38, R) >droAna3.scaffold_13337 4958877 142 - 23293914 ----GCUGGUACUGGUGUUAUUGUCUACGUGGCAGAACUUCCGCUGCCACUGCCACUGUGGUUGGCUGCUGCGGUGGAUUUGCCGGCUUUGGUGGCUUUGGCGGCUGCGGCAACCGGAAGUGCCAAACACUUUCCGGUCACGCAGU ----.........((.(((.(((((.....))))))))..))(((((((..(((((((.(((((((..((.....))....))))))).)))))))..)))))))((((...((((((((((.....))).)))))))..)))).. ( -63.10, z-score = -1.85, R) >dp4.chrXR_group8 7185436 105 + 9212921 ---GGCUGCUAGUGGC--CAUUGUCUACGUGGCAGGACUUCCGCUGC---------UGUUGUCGGCUGCUGCGGUGG---------------------------CUGCGGCAACCGGAAACGCCAAACACUUUCCGGUCACGCAGU ---.(((((.((..((--(...(.(.(((..((.((....))))..)---------.)).).))))..))))))).(---------------------------(((((...((((((((..........))))))))..)))))) ( -44.50, z-score = -1.06, R) >droPer1.super_36 697673 105 - 818889 ---GGCUGCUAGUGGC--CAUUGUCUACGUGGCAGGACUUCCGCUGC---------UGUUGUCGGCUGCUGCGGUGG---------------------------CUGCGGCAACCGGAAACGCCAAACACUUUCCGGUCACGCAGU ---.(((((.((..((--(...(.(.(((..((.((....))))..)---------.)).).))))..))))))).(---------------------------(((((...((((((((..........))))))))..)))))) ( -44.50, z-score = -1.06, R) >droWil1.scaffold_180949 3211579 113 - 6375548 ------------UGUUAUUAUUGUCUACGCGGCAAAACUUCCGCCAGC--------UGUUGUUG-CUAUUGCUAUUGCU------------GAUGGUGCGGUGGCUGUGGCAACCGGAAAUGCCAAACACUUUCCGGUCACGCAGU ------------................((((........))))..((--------(((...((-((((.((((((((.------------......)))))))).))))))((((((((((.....)).))))))))...))))) ( -38.30, z-score = -1.19, R) >droGri2.scaffold_15110 5819372 139 - 24565398 UAUUGUAGUCGCUGUUGUUGUUGUCUACGUGGCAAA-CUUCCGUUUGAUG------UUGCCUCGGUGGCUUGAGGAAUUGUAACCACGACUCAUUCGUCGGUGGCUGCGGUAACCGGAAACGCGAAACACUUUCCGGUCACGCAGU ((((((((((((((.((.((..(((..((.(((((.-..((.....))..------))))).))((((.(((........)))))))))).))..)).))))))))))))))((((((((..........))))))))........ ( -45.80, z-score = -0.85, R) >droVir3.scaffold_13049 11919673 133 - 25233164 GGUUGUUUUUGUUGUUGCUGCUGUCCAGGCGGUUAG-CUUCCGUUCGGUG------UGGCCGCUGUUGUUGGCAGCG------CCGCAACUCAUUCGUUGGUGGCUGCGUCAACCGGAAACGCCAAACACUUUCCGGUCACGCAGU ..........(((((.((.((((((..((((((((.-(..((....)).)------))))))))......)))))))------).))))).(((......)))(((((((..((((((((..........)))))))).))))))) ( -49.70, z-score = -0.68, R) >consensus ____GCUUGUAUUGGUGUUAUUGUCUACGUGGCAAAACUUCCGCUGC_________UGGUGUUGGCUGCUGCGGUGGUUUUG_________GCCGCUUCGGUGGCUGCGGCAACCGGAAACGCCAAACACUUUCCGGUCACGCAGU ...........................(((((((................................)))))))..............................((((((...((((((((..........))))))))..)))))) (-22.57 = -22.55 + -0.02)
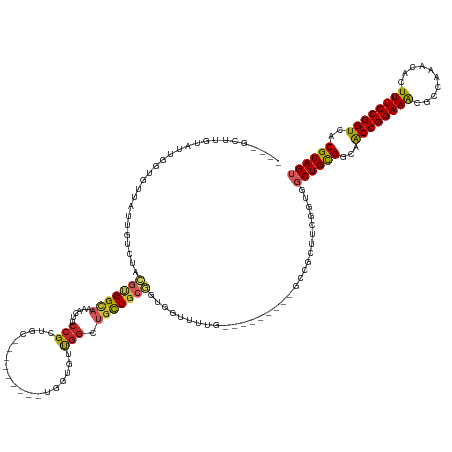
| Location | 19,118,066 – 19,118,189 |
|---|---|
| Length | 123 |
| Sequences | 10 |
| Columns | 146 |
| Reading direction | reverse |
| Mean pairwise identity | 69.01 |
| Shannon entropy | 0.61562 |
| G+C content | 0.56927 |
| Mean single sequence MFE | -42.06 |
| Consensus MFE | -21.77 |
| Energy contribution | -21.72 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.987545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19118066 123 - 24543557 ACUGCGUGACCGGAAAGUGUUUGCUGCUUCCGGUUGCCGCAGCCACCGAAGCGGC---------CAAAACCACCGCAGCAGCCAAAACC----------GCAGCGGAAGUUUUGCCACGUAGACAAUAACACCCAUACAACC---- .(((((((((((((((((....)))..))))))).(((((..........)))))---------((((((..((((.((.(......).----------)).))))..)))))).)))))))....................---- ( -42.10, z-score = -2.66, R) >droSim1.chr3L 18424409 123 - 22553184 ACUGCGUGACCGGAAAGUGUUUGCUGCUUCCGGUUGCCGCAGCCACCGAAGCGGC---------CAAAACCACCGCAGCAGCCAAAACC----------GCAGCGGAAGUUUUGCCACGUAGACAAUAACACCCAUACAACC---- .(((((((((((((((((....)))..))))))).(((((..........)))))---------((((((..((((.((.(......).----------)).))))..)))))).)))))))....................---- ( -42.10, z-score = -2.66, R) >droYak2.chr3L 2726811 123 + 24197627 ACUGCGUGACCGGAAAGUGUUUGGUACUUCCGGCUGCCGCAGCCACCGAAGCGGC---------CAAAACCACCGCAGCAGCCAAAACC----------GCAGCGGAAGUUUUGCCACGUAGACAAUAACACCCAUACAAGC---- .(((((((.((((((.(((.....)))))))))..(((((..........)))))---------((((((..((((.((.(......).----------)).))))..)))))).)))))))....................---- ( -41.20, z-score = -2.10, R) >droEre2.scaffold_4784 18929875 123 - 25762168 ACUGCGUGACCGGAAAGUGUUUGGUGCUUCCGGCUGCCGCAGCCACCGAAGCGGC---------CAAAACCACCGCAGCAGCCAAAACC----------GCAGCGGAAGUUUUGCCACGUAGACAAUAACACCCAUACAAUC---- .((((((.((((((.....))))))(((((.(((((...)))))...)))))(((---------.(((((..((((.((.(......).----------)).))))..))))))))))))))....................---- ( -41.60, z-score = -2.06, R) >droAna3.scaffold_13337 4958877 142 + 23293914 ACUGCGUGACCGGAAAGUGUUUGGCACUUCCGGUUGCCGCAGCCGCCAAAGCCACCAAAGCCGGCAAAUCCACCGCAGCAGCCAACCACAGUGGCAGUGGCAGCGGAAGUUCUGCCACGUAGACAAUAACACCAGUACCAGC---- .((((((((((((((.(((.....))))))))))((((((.(((((....(((.........))).........((....))........))))).))))))((((.....)))))))))))....................---- ( -51.60, z-score = -1.70, R) >dp4.chrXR_group8 7185436 105 - 9212921 ACUGCGUGACCGGAAAGUGUUUGGCGUUUCCGGUUGCCGCAG---------------------------CCACCGCAGCAGCCGACAACA---------GCAGCGGAAGUCCUGCCACGUAGACAAUG--GCCACUAGCAGCC--- .(((((..((((((((.((.....))))))))))..)....(---------------------------(((((((.((...........---------)).))))..(((.(((...))))))..))--)).....))))..--- ( -37.80, z-score = -0.51, R) >droPer1.super_36 697673 105 + 818889 ACUGCGUGACCGGAAAGUGUUUGGCGUUUCCGGUUGCCGCAG---------------------------CCACCGCAGCAGCCGACAACA---------GCAGCGGAAGUCCUGCCACGUAGACAAUG--GCCACUAGCAGCC--- .(((((..((((((((.((.....))))))))))..)....(---------------------------(((((((.((...........---------)).))))..(((.(((...))))))..))--)).....))))..--- ( -37.80, z-score = -0.51, R) >droWil1.scaffold_180949 3211579 113 + 6375548 ACUGCGUGACCGGAAAGUGUUUGGCAUUUCCGGUUGCCACAGCCACCGCACCAUC------------AGCAAUAGCAAUAG-CAACAACA--------GCUGGCGGAAGUUUUGCCGCGUAGACAAUAAUAACA------------ .(((((..((((((((.((.....))))))))))..)..((((....((......------------.))....((....)-).......--------))))((((........))))))))............------------ ( -35.10, z-score = -1.40, R) >droGri2.scaffold_15110 5819372 139 + 24565398 ACUGCGUGACCGGAAAGUGUUUCGCGUUUCCGGUUACCGCAGCCACCGACGAAUGAGUCGUGGUUACAAUUCCUCAAGCCACCGAGGCAA------CAUCAAACGGAAG-UUUGCCACGUAGACAACAACAACAGCGACUACAAUA .((((((((((((((((((...))).)))))))))).))))).....(((......)))(((((..(..........(((.....)))..------...(((((....)-))))....................)..))))).... ( -45.30, z-score = -2.72, R) >droVir3.scaffold_13049 11919673 133 + 25233164 ACUGCGUGACCGGAAAGUGUUUGGCGUUUCCGGUUGACGCAGCCACCAACGAAUGAGUUGCGG------CGCUGCCAACAACAGCGGCCA------CACCGAACGGAAG-CUAACCGCCUGGACAGCAGCAACAACAAAAACAACC .(((((((((((((((.((.....))))))))))).))))))..............(((((((------(((((.......)))).))).------..(((..(((...-....)))..)))...)))))................ ( -46.00, z-score = -1.49, R) >consensus ACUGCGUGACCGGAAAGUGUUUGGCGCUUCCGGUUGCCGCAGCCACCGAAGCGGC_________CAAAACCACCGCAGCAGCCAAAACCA_________GCAGCGGAAGUUUUGCCACGUAGACAAUAACACCAAUACAAGC____ .(((((((...((((.(((.....)))))))((((((.((..................................)).))))))...................((....)).....)))))))........................ (-21.77 = -21.72 + -0.05)
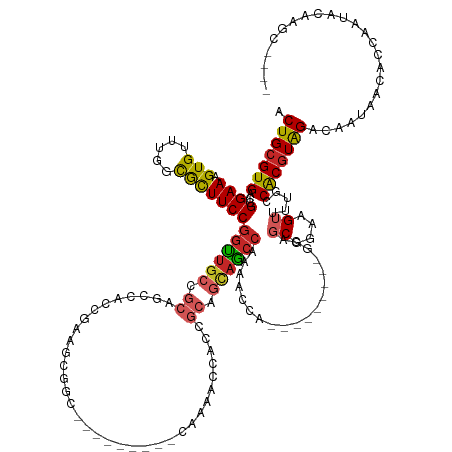
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:10 2011