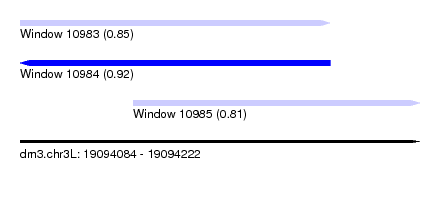
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,094,084 – 19,094,222 |
| Length | 138 |
| Max. P | 0.924316 |

| Location | 19,094,084 – 19,094,191 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.21 |
| Shannon entropy | 0.16393 |
| G+C content | 0.45704 |
| Mean single sequence MFE | -25.64 |
| Consensus MFE | -21.13 |
| Energy contribution | -20.97 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
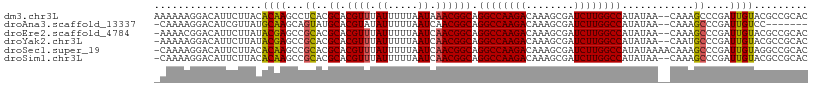
>dm3.chr3L 19094084 107 + 24543557 AAAAAAGGACAUUCUUACACAAGCCUCACGCACGUUUAUUUUUAAUAAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAA--CAAAGCCCGAUUGUACGCCGCAC ......(((((.((...............((.(((((((.....))))))))).((((((((........))))))))......--........)).)))...)).... ( -26.20, z-score = -2.78, R) >droAna3.scaffold_13337 4941831 99 - 23293914 -CAAAAGGACAUCGUUAUGCAAGCAGUAUGCACGUAUAUUUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAA--CAAAGCCCGAUUGUCC------- -.....((((((((....((........(((.(((.............))))))((((((((........))))))))......--....)).))).)))))------- ( -27.52, z-score = -3.09, R) >droEre2.scaffold_4784 18913179 106 + 25762168 -AAAACGGACAUUCUUAUACGAGCCGCACGCACGUUUAUUUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAA--CAAAGCCCGAUUGUACGCCGCAC -.(((((.....((......))((.....)).)))))............((((.((((((((........)))))))).....(--(((.......))))..))))... ( -25.00, z-score = -1.44, R) >droYak2.chr3L 2709934 106 - 24197627 -AAAAAGGACAUUCUUAUACGAGCCGCACGCACGUUUAUUUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAA--CAAUGCCCGAUUGUACGCCGCAC -..............((.(((.((.....)).))).))...........((((.((((((((........)))))))).....(--((((.....)))))..))))... ( -26.20, z-score = -1.76, R) >droSec1.super_19 1056868 108 + 1257711 -CAAAAGGACAUUCUUACACAAGCCGCACGCACGUUUAUUUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAAAACAAAGCCCGAUUGUAGGCCGCAC -.....((.....((((((......((..((.((((.((.....)).)))))).((((((((........))))))))............)).....)))))))).... ( -24.80, z-score = -1.56, R) >droSim1.chr3L 18407541 106 + 22553184 -CAAAAGGACAUUCUUACACAAGCCGCACGCACGUUUAUUUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAA--CAAAGCCCGAUUGUACGCCGCAC -.....................((.((..((.((((.((.....)).)))))).((((((((........))))))))......--................)).)).. ( -24.10, z-score = -1.69, R) >consensus _AAAAAGGACAUUCUUACACAAGCCGCACGCACGUUUAUUUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAA__CAAAGCCCGAUUGUACGCCGCAC ..................((((...((..((.((((.((.....)).)))))).((((((((........))))))))............))....))))......... (-21.13 = -20.97 + -0.16)
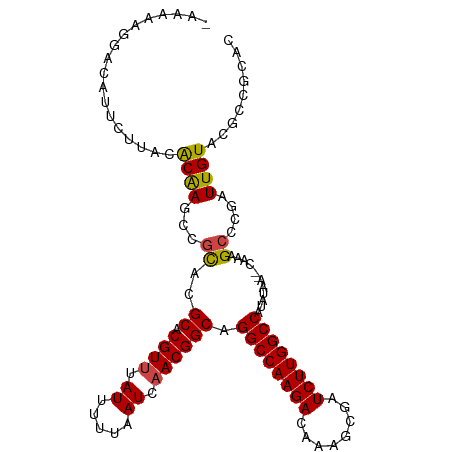
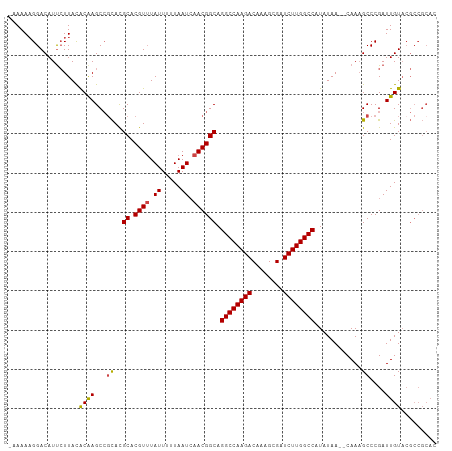
| Location | 19,094,084 – 19,094,191 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.21 |
| Shannon entropy | 0.16393 |
| G+C content | 0.45704 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -26.34 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19094084 107 - 24543557 GUGCGGCGUACAAUCGGGCUUUG--UUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUUAUUAAAAAUAAACGUGCGUGAGGCUUGUGUAAGAAUGUCCUUUUUU (..((...((((..(((((((..--(.....(((((((((......))))))))).(((((((((.....))))))).)))..)))))))))))....))..)...... ( -35.60, z-score = -2.94, R) >droAna3.scaffold_13337 4941831 99 + 23293914 -------GGACAAUCGGGCUUUG--UUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUGAUUAAAAAUAUACGUGCAUACUGCUUGCAUAACGAUGUCCUUUUG- -------(((((.((((((....--......(((((((((......))))))))).))).................(((((.......)))))..)))))))).....- ( -32.90, z-score = -3.75, R) >droEre2.scaffold_4784 18913179 106 - 25762168 GUGCGGCGUACAAUCGGGCUUUG--UUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUGAUUAAAAAUAAACGUGCGUGCGGCUCGUAUAAGAAUGUCCGUUUU- ((((...))))...(((((.((.--(((((((((((((((......))))))))).(((((.....................)))))..)))))).)).)))))....- ( -35.20, z-score = -1.99, R) >droYak2.chr3L 2709934 106 + 24197627 GUGCGGCGUACAAUCGGGCAUUG--UUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUGAUUAAAAAUAAACGUGCGUGCGGCUCGUAUAAGAAUGUCCUUUUU- ((((...))))....(((((((.--(((((((((((((((......))))))))).(((((.....................)))))..)))))).))))))).....- ( -37.90, z-score = -3.04, R) >droSec1.super_19 1056868 108 - 1257711 GUGCGGCCUACAAUCGGGCUUUGUUUUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUGAUUAAAAAUAAACGUGCGUGCGGCUUGUGUAAGAAUGUCCUUUUG- ..........(((..((((....(((((((((((((((((......))))))))).(((((.....................)))))..))))))))..))))..)))- ( -32.60, z-score = -1.75, R) >droSim1.chr3L 18407541 106 - 22553184 GUGCGGCGUACAAUCGGGCUUUG--UUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUGAUUAAAAAUAAACGUGCGUGCGGCUUGUGUAAGAAUGUCCUUUUG- ((((...))))....((((.((.--(((((((((((((((......))))))))).(((((.....................)))))..)))))).)).)))).....- ( -33.50, z-score = -1.82, R) >consensus GUGCGGCGUACAAUCGGGCUUUG__UUAUAUGGCCAAGAUCGCUUUGUCUUGGCCUGCCGUUGAUUAAAAAUAAACGUGCGUGCGGCUUGUAUAAGAAUGUCCUUUUG_ ...............((((......(((((((((((((((......)))))))))(((((((...........)))).)))........))))))....))))...... (-26.34 = -26.28 + -0.05)

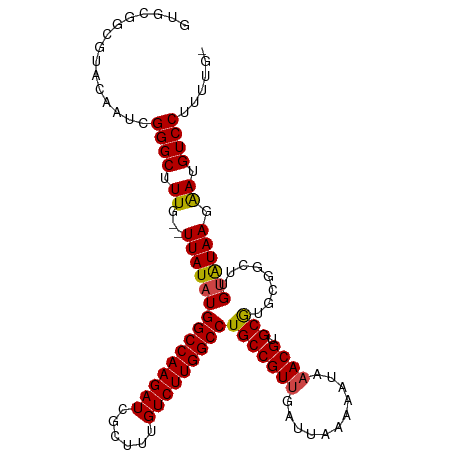
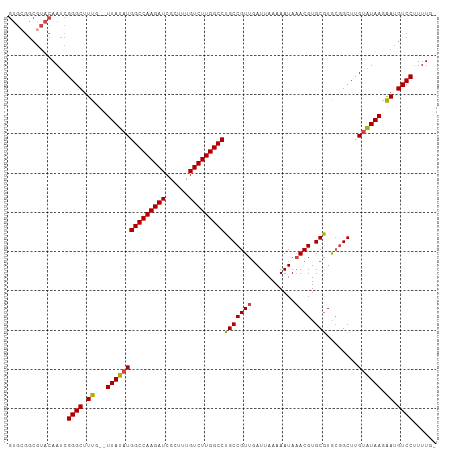
| Location | 19,094,123 – 19,094,222 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 84.00 |
| Shannon entropy | 0.27468 |
| G+C content | 0.51510 |
| Mean single sequence MFE | -26.56 |
| Consensus MFE | -20.34 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.810997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19094123 99 + 24543557 UUUUAAUAAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUA--ACAAAGCCCGAUUGUACGCCGCACUUAUCCGGCAGCACGAUGUCCUUCUGCGAGU ............(((((((((((........)))))))).....--...(((..(.(((((.(((((........)))).).))))))..))).))).... ( -25.80, z-score = -0.83, R) >droAna3.scaffold_13337 4941869 88 - 23293914 UUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUA--ACAAAGCCCGAUUGU---CCCUGCCUGGCUU-CCGUCCUUUGUCGCCA------- ...........(((.((((((((........)))))))).....--((((((..((...((---(.......)))..-.))..)))))).))).------- ( -28.10, z-score = -2.92, R) >droEre2.scaffold_4784 18913217 90 + 25762168 UUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUA--ACAAAGCCCGAUUGUACGCCGCACUUACCCGGCAGCCCGAUGUCCU--------- .....(((...(((.((((((((........)))))))).....--.................((((........)))).))).))).....--------- ( -25.40, z-score = -1.97, R) >droYak2.chr3L 2709972 90 - 24197627 UUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUA--ACAAUGCCCGAUUGUACGCCGCACUUACCCGGCAGCCCGAUGUCCU--------- .....(((...(((.((((((((........)))))))).....--(((((.....)))))..((((........)))).))).))).....--------- ( -26.90, z-score = -2.34, R) >droSec1.super_19 1056906 101 + 1257711 UUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUAAAACAAAGCCCGAUUGUAGGCCGCACUUACCCGGCAGCCCGAUGUCCUCCUGCGAGU .....(((...(((.((((((((........))))))))............(((.(...(((((.....)))))).))).))).)))...(((....))). ( -26.60, z-score = -0.65, R) >consensus UUUUAAUCAACGGCAGGCCAAGACAAAGCGAUCUUGGCCAUAUA__ACAAAGCCCGAUUGUACGCCGCACUUACCCGGCAGCCCGAUGUCCU_________ ...........(((.((((((((........))))))))........................((((........)))).))).................. (-20.34 = -21.18 + 0.84)

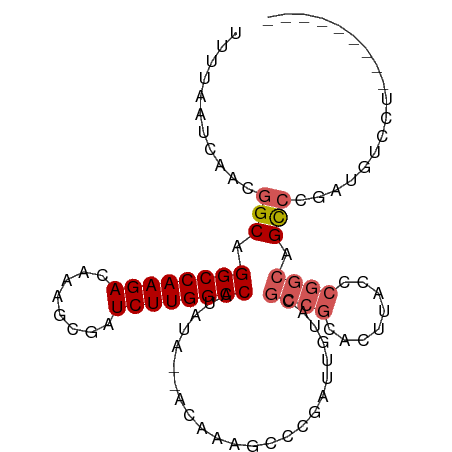
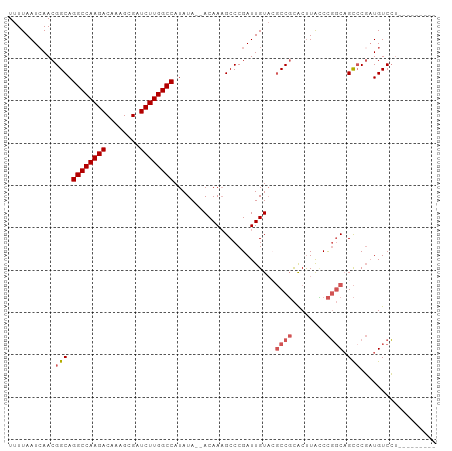
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:08 2011