| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,081,374 – 19,081,482 |
| Length | 108 |
| Max. P | 0.991008 |

| Location | 19,081,374 – 19,081,482 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.24 |
| Shannon entropy | 0.63906 |
| G+C content | 0.55599 |
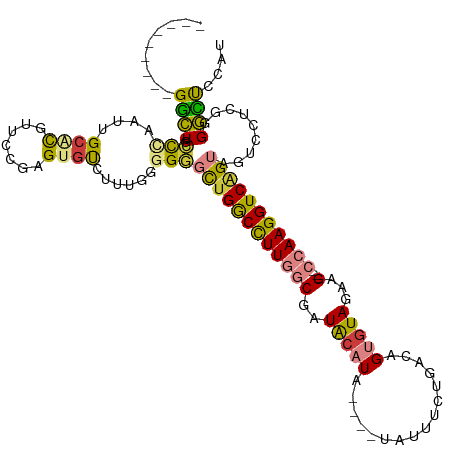
| Mean single sequence MFE | -41.79 |
| Consensus MFE | -17.11 |
| Energy contribution | -16.33 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.991008 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
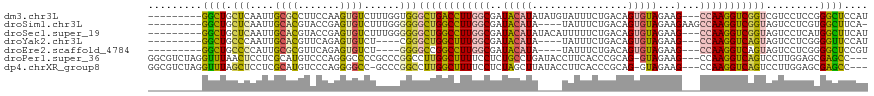
>dm3.chr3L 19081374 108 + 24543557 ---------GGCUGCUCAAUUGCGCCUUCCAAGUGUCUUUGGUGGGCUGACCUUGGCGAUACAUAUAUGUAUUUCUGACAGUGUAGAAG---CCAAGGUCGGUCGUCCUCCGGGCUCCAU ---------(((.((......)))))......(.((((..((..(((((((((((((..(((((...(((.......))))))))...)---))))))))))))..))...)))).)... ( -42.00, z-score = -2.82, R) >droSim1.chr3L 18394794 106 + 22553184 ---------GGCUGCUCAAUUGCACGUACCGAGUGUCUUUGGGGGGCUGGCCUUGGCGAUACAUA----UAUUUCUGACAGUGUAGAAGAAGCCAAGGUCGGUAGUCCUCGUGGCUUCA- ---------(((..(.(((..((((.......))))..)))((((((((((((((((......((----((((......))))))......)))))))))...))))))))..)))...- ( -41.80, z-score = -2.50, R) >droSec1.super_19 1044856 108 + 1257711 ---------GGCUGCUCAAUUGCACGUACCGAGUGUCUUUGGGGGGCUGGCCUUGGCGAUACAUAUACAUUUUUCUGACAGUGUAGAAG---CCAAGGUCGGUAGUCCUCAUGGCUUCAU ---------.(((((......))).))...(((.(((..((((((((((((((((((..(((((...((......))...)))))...)---)))))))))))..)))))).)))))).. ( -41.10, z-score = -2.65, R) >droYak2.chr3L 2697057 100 - 24197627 ---------GGCUGCCCAAUUGCACGUUCAGAGUGUCU----CGGGCUGGCUUUGGCGAUACAUA----UAUUUCUGACAGUGUAGAAG---CCAAGGUCAGUAGUCCUCGGGGUUCCAU ---------((..((((....((((.......))))..----.((((((((((((((..(((((.----...........)))))...)---)))))))))))...))...)))).)).. ( -36.20, z-score = -2.10, R) >droEre2.scaffold_4784 18900242 100 + 25762168 ---------GGCUGCCCCAUUGCGCGUUCAGAGUGUCU----GGGGCCGGCCUUGGCGAUACAUA----UAUUUCUGACAGUGUAGAAG---CCAAGGUCAGUAGUCCUCGGGGCUCCGU ---------((..((((((((((((.(((((.....))----))))).(((((((((..(((((.----...........)))))...)---)))))))).)))))....))))).)).. ( -42.10, z-score = -2.27, R) >droPer1.super_36 671093 113 - 818889 GGCGUCUAGGUUUAACUCCUCGCAUGUCCCAGGGCCCCGCCCGGCCUUGGCUUUUCCUCUGCCUGAUACCUUCACCCGCAG-GUAGAAG---CCAAGGUCAGUCCUUGGAGCGAGCC--- (((.....((.......))((((...(((..((((...))))(((((((((((((.(.((((.(((.....)))...))))-).)))))---)))))))).......))))))))))--- ( -45.90, z-score = -2.41, R) >dp4.chrXR_group8 7158886 112 + 9212921 GGCGUCUAGGUUUAGCUCCUCGCAUGUCCCAGGGGCC-GCCCGGCCUUGGCUUUUCCUCUAGCUUAUACCUUCACCCGCAG-GUAGAAG---CCAAGGUCAGUCCUUGGAGCGAGCC--- (((((..(((.......))).)).((..((((((((.-....((((((((((((((((...((..............))))-).)))))---)))))))).))))))))..)).)))--- ( -43.44, z-score = -1.34, R) >consensus _________GGCUGCCCAAUUGCACGUUCCGAGUGUCUUUGGGGGGCUGGCCUUGGCGAUACAUA____UAUUUCUGACAGUGUAGAAG___CCAAGGUCAGUAGUCCUCGGGGCUCCAU .........((((.(((....((((.......))))......)))(((((((((((...(((((................))))).......))))))))))).........)))).... (-17.11 = -16.33 + -0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:05 2011