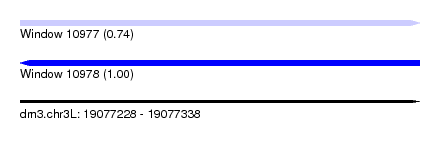
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,077,228 – 19,077,338 |
| Length | 110 |
| Max. P | 0.996917 |

| Location | 19,077,228 – 19,077,338 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.56321 |
| G+C content | 0.34318 |
| Mean single sequence MFE | -21.17 |
| Consensus MFE | -10.97 |
| Energy contribution | -12.42 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
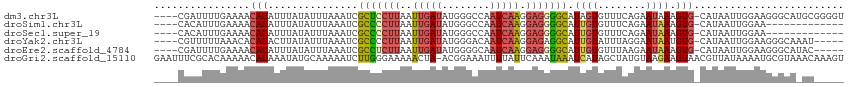
>dm3.chr3L 19077228 110 + 24543557 ----CGAUUUUGAAAACACAUUUAUAUUUAAAUCGCUCCUUAAUUGAUAUGGGCCAAUCAAGGAGGGGCAUAGUGUUUCAGAAUAAAGUG-CAUAAUUGGAAGGGCAUGCGGGGU ----..(((((((((.((((((((....))))).(((((((..(((((........))))).)))))))...))))))))))))....((-(((..((....))..))))).... ( -27.00, z-score = -1.70, R) >droSim1.chr3L 18390437 97 + 22553184 ----CACAUUUGAAAACACAUUUAUAUUUAAAUCGCCCCUUAAUUGAUAUGGGCCAAUCAAGGAGGGGCAUUGUGUUUCAGAAUAAAGUG-CAUAAUUGGAA------------- ----....(((((((.((((((((....))))..(((((((..(((((........))))).)))))))..)))))))))))........-...........------------- ( -25.20, z-score = -2.14, R) >droSec1.super_19 1040627 97 + 1257711 ----CACAUUUGAAAACACAUUUAUAUUUAAAUCGCCCCUUAAUUGAUAUGGGCCAAUCAAGGAGGGGCAUUGUGUUUCAGAAUAAAGUG-CAUAAUUGGAA------------- ----....(((((((.((((((((....))))..(((((((..(((((........))))).)))))))..)))))))))))........-...........------------- ( -25.20, z-score = -2.14, R) >droYak2.chr3L 2692629 105 - 24197627 ----CGUUUUUAAACACACACUUAUAUUUAAAUCGCCCCUUAAUUGAUAUGGGACAAUCAAGGAGAGGCAUUGUAUUUAGGAAUAAUGUG-CAUAAUUGGAAGGGCAAAU----- ----.((((((...((((((.((((.(((((((((((.(((..(((((........))))).))).)))...).))))))).))))))))-......)).))))))....----- ( -17.00, z-score = 0.33, R) >droEre2.scaffold_4784 18896025 105 + 25762168 ----CGAUUUUGAAAACACAUUUAUAUUUAAAUCGCCUCUUAAUUGAUAUGGGGCAAUCAAGGAGGGGCAUUGUGUUUAAGAAUAAAGUG-CAUAAUUGGAAGGGCAUAC----- ----..(((((..(((((((((((....))))..(((((((..(((((........))))).)))))))..))))))).)))))...(((-(............))))..----- ( -23.10, z-score = -2.11, R) >droGri2.scaffold_15110 5783925 114 - 24565398 GAAUUUCGCACAAAAACACAAAUAUGCAAAAAUCUUGGGAAAAACUA-ACGGAAAUUUUAUUCAAAUAAAUCAUAGCUAUGUAAGAAGUAACGUUAUAAAAUGCGUAAACAAAGU ......................((((((....((....)).....((-((((((......)))............(((........)))..))))).....))))))........ ( -9.50, z-score = 1.00, R) >consensus ____CGCUUUUGAAAACACAUUUAUAUUUAAAUCGCCCCUUAAUUGAUAUGGGCCAAUCAAGGAGGGGCAUUGUGUUUAAGAAUAAAGUG_CAUAAUUGGAAGGGCA_AC_____ ........(((..(((((((((((....))))).(((((((..(((((........))))).)))))))...))))))..)))................................ (-10.97 = -12.42 + 1.45)

| Location | 19,077,228 – 19,077,338 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.46 |
| Shannon entropy | 0.56321 |
| G+C content | 0.34318 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -12.14 |
| Energy contribution | -11.65 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
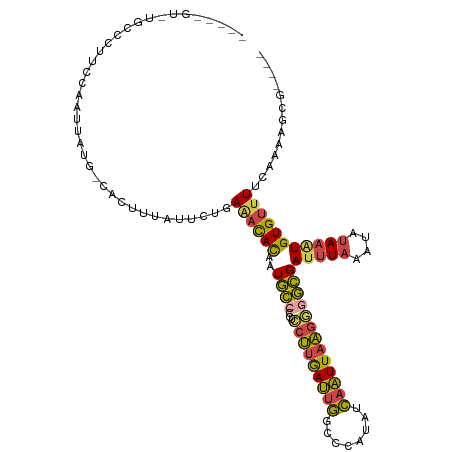
>dm3.chr3L 19077228 110 - 24543557 ACCCCGCAUGCCCUUCCAAUUAUG-CACUUUAUUCUGAAACACUAUGCCCCUCCUUGAUUGGCCCAUAUCAAUUAAGGAGCGAUUUAAAUAUAAAUGUGUUUUCAAAAUCG---- .....(((((..........))))-)..........(((((((......(((((((((((((......)))))))))))).)(((((....))))))))))))........---- ( -23.10, z-score = -3.48, R) >droSim1.chr3L 18390437 97 - 22553184 -------------UUCCAAUUAUG-CACUUUAUUCUGAAACACAAUGCCCCUCCUUGAUUGGCCCAUAUCAAUUAAGGGGCGAUUUAAAUAUAAAUGUGUUUUCAAAUGUG---- -------------...........-.....((((..(((((((..(((((((...(((((((......))))))))))))))(((((....))))))))))))..))))..---- ( -23.30, z-score = -2.58, R) >droSec1.super_19 1040627 97 - 1257711 -------------UUCCAAUUAUG-CACUUUAUUCUGAAACACAAUGCCCCUCCUUGAUUGGCCCAUAUCAAUUAAGGGGCGAUUUAAAUAUAAAUGUGUUUUCAAAUGUG---- -------------...........-.....((((..(((((((..(((((((...(((((((......))))))))))))))(((((....))))))))))))..))))..---- ( -23.30, z-score = -2.58, R) >droYak2.chr3L 2692629 105 + 24197627 -----AUUUGCCCUUCCAAUUAUG-CACAUUAUUCCUAAAUACAAUGCCUCUCCUUGAUUGUCCCAUAUCAAUUAAGGGGCGAUUUAAAUAUAAGUGUGUGUUUAAAAACG---- -----..((((((..........(-((...((((....))))...))).....((((((((........))))))))))))))((((((((((....))))))))))....---- ( -22.30, z-score = -2.82, R) >droEre2.scaffold_4784 18896025 105 - 25762168 -----GUAUGCCCUUCCAAUUAUG-CACUUUAUUCUUAAACACAAUGCCCCUCCUUGAUUGCCCCAUAUCAAUUAAGAGGCGAUUUAAAUAUAAAUGUGUUUUCAAAAUCG---- -----(((((..........))))-)...........((((((..((((....((((((((........)))))))).))))(((((....))))))))))).........---- ( -15.40, z-score = -1.79, R) >droGri2.scaffold_15110 5783925 114 + 24565398 ACUUUGUUUACGCAUUUUAUAACGUUACUUCUUACAUAGCUAUGAUUUAUUUGAAUAAAAUUUCCGU-UAGUUUUUCCCAAGAUUUUUGCAUAUUUGUGUUUUUGUGCGAAAUUC ..........(((((..................((((((.((((....((((....((((((.....-.)))))).....)))).....)))).))))))....)))))...... ( -13.45, z-score = 0.13, R) >consensus _____GU_UGCCCUUCCAAUUAUG_CACUUUAUUCUGAAACACAAUGCCCCUCCUUGAUUGGCCCAUAUCAAUUAAGGGGCGAUUUAAAUAUAAAUGUGUUUUCAAAAGCG____ .....................................((((((..((((...(((((((((........)))))))))))))(((((....)))))))))))............. (-12.14 = -11.65 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:37:03 2011