| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,037,884 – 19,037,979 |
| Length | 95 |
| Max. P | 0.912436 |

| Location | 19,037,884 – 19,037,979 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.35 |
| Shannon entropy | 0.41120 |
| G+C content | 0.38708 |
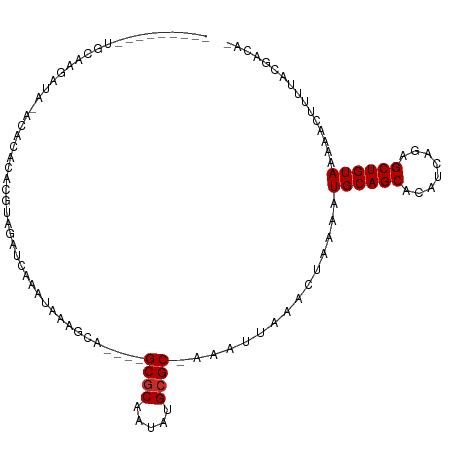
| Mean single sequence MFE | -20.26 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.912436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
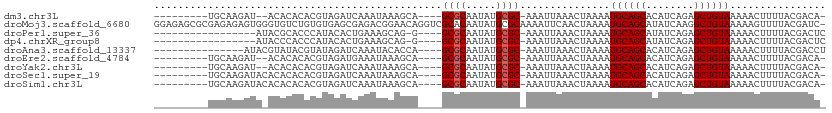
>dm3.chr3L 19037884 95 - 24543557 ---------UGCAAGAU--ACACACACGUAGAUCAAAUAAAGCA----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACA- ---------........--.......((((((............----((((.....))))-..............((((((........))))))......))))))...- ( -20.00, z-score = -2.18, R) >droMoj3.scaffold_6680 13959459 111 - 24764193 GGAGAGCGCGAGAGAGUGGGUGUCUGUGUGAGCGAGACGGAACAGGUCGCACAAUAUGCGCAAAAUUCAACUAAAAUGCAGCAUAUCAAGGCUGUAAAAAGUUUUACGAUC- ..(((((........(((.((((.(((((((.(...........).))))))))))).)))...............((((((........))))))....)))))......- ( -27.40, z-score = -0.40, R) >droPer1.super_36 625858 89 + 818889 -----------------AUACGCACCCAUACACUGAAAGCAG-G----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCAUAUCAGAGCUGUAAAAACUUUUACGACUC -----------------....((.(((..............)-)----).)).((((((((-(.............))).))))))..(((.((((((....)))))).))) ( -18.56, z-score = -1.65, R) >dp4.chrXR_group8 7113764 89 - 9212921 -----------------AUACCCACCCAUACACUGAAAGCAG-G----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCAUAUCAGAGCUGUAAAAACUUUUACGACUC -----------------................((((((...-.----((((.....))))-..............((((((........))))))....))))))...... ( -17.30, z-score = -1.66, R) >droAna3.scaffold_13337 4890118 92 + 23293914 ---------------AUACGUAUACGUAUAGAUCAAAUACACCA----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACCU ---------------...((((...((((.......))))....----((((.....))))-..............((((((........))))))........)))).... ( -18.90, z-score = -2.63, R) >droEre2.scaffold_4784 18856286 95 - 25762168 ---------UGCAAGAU--ACACACACGUAGAUGAAAUAAAGCA----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACA- ---------........--.......((((((.(..........----((((.....))))-..............((((((........))))))....).))))))...- ( -20.20, z-score = -1.98, R) >droYak2.chr3L 2638952 95 + 24197627 ---------UGCAAGAU--ACACACACGUAGAUCAAAUAAAGCA----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACA- ---------........--.......((((((............----((((.....))))-..............((((((........))))))......))))))...- ( -20.00, z-score = -2.18, R) >droSec1.super_19 1001897 97 - 1257711 ---------UGCAAGAUACACACACACGUAGAUCAAAUAAAGCA----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACA- ---------.................((((((............----((((.....))))-..............((((((........))))))......))))))...- ( -20.00, z-score = -2.20, R) >droSim1.chr3L 18347687 97 - 22553184 ---------UGCAAGAUACACACACACGUAGAUCAAAUAAAGCA----GCGCAAUAUGCGC-AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACA- ---------.................((((((............----((((.....))))-..............((((((........))))))......))))))...- ( -20.00, z-score = -2.20, R) >consensus _________UGCAAGAUA_ACACACACGUAGAUCAAAUAAAGCA____GCGCAAUAUGCGC_AAAUUAAACUAAAAUGCAGCACAUCAGAGCUGUAAAAACUUUUACGACA_ ................................................((((.....))))...............((((((........))))))................ (-12.46 = -12.57 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:56 2011