
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,026,251 – 19,026,354 |
| Length | 103 |
| Max. P | 0.652327 |

| Location | 19,026,251 – 19,026,348 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Shannon entropy | 0.34233 |
| G+C content | 0.51892 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.652327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
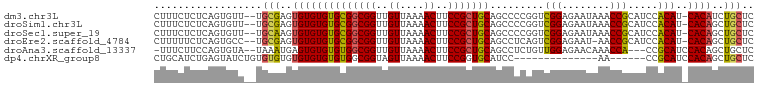
>dm3.chr3L 19026251 97 - 24543557 CUCAGUGUU--UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU-CACAUCUGCUCCAAAAU .......((--((.(((((((((((((((((..((....))..)))))))).....((.(((........)))...))....-)))))..)))))))).. ( -30.40, z-score = -1.61, R) >droSim1.chr3L 18335071 97 - 22553184 CUCAGUGUU--UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU-CACAGCUGCUCCAAAAU ....(((..--(((((.((.(..((((((((..((....))..))))))))..).)).))((........)))))..)))..-................. ( -29.40, z-score = -0.80, R) >droSec1.super_19 990366 97 - 1257711 CUCAGUGUU--UGCAAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU-CACAGCUGCUCCAAAAU .........--.(((..((((((((..((((((........((((((((......))).)))))....))))))...)))).-))))..)))........ ( -29.10, z-score = -0.98, R) >droEre2.scaffold_4784 18844454 96 - 25762168 CUCAGUGCC--UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCUCAGUCGGAGAAU-AACCGCAUCCACAU-CACAGCUGCUCCAAAAU ....(.((.--.((..(((.(((((..(((((((((.....((((((((......))).))))))))-))))))...)))))-))).)).)).)...... ( -32.50, z-score = -1.89, R) >droAna3.scaffold_13337 4878040 95 + 23293914 UCCAGUGUA--UAAAUGAGUGUGUGUGGCGGUUGUUAAAACUUCCGCUGCAGCCUCUGUUGGAGAACAAACCA---CCGCAUCCACAGCUGCUCCAAAAA ..((((...--.......(((..(((((.(((((((.....(((((..((((...)))))))))))).)))).---)))))..))).))))......... ( -24.40, z-score = -0.65, R) >dp4.chrXR_group8 7101760 80 - 9212921 CUGAGUAUCUGUGUGUGUGUGUGUGUGGCGGUAGUUAAAACUUCCGGUGCAUCC----------AA----------CCGCAUCCACAGCUGCUCCAAAAU ..(((((.(((((..((((((..((((.(((.(((....))).))).))))..)----------).----------.))))..))))).)))))...... ( -30.90, z-score = -4.37, R) >consensus CUCAGUGUU__UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU_CACAGCUGCUCCAAAAU ..((((((....))..(((.(((((((((((..((....))..))))))).........(((........))).....)))).))).))))......... (-18.64 = -18.43 + -0.21)


| Location | 19,026,257 – 19,026,354 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Shannon entropy | 0.36784 |
| G+C content | 0.52910 |
| Mean single sequence MFE | -29.25 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.555162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19026257 97 - 24543557 CUUUCUCUCAGUGUU--UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU-CACAUCUGCUC ...............--.(((..((((((((..((((((........((((((((......))).)))))....))))))...)))).-))))..))).. ( -30.10, z-score = -1.51, R) >droSim1.chr3L 18335077 97 - 22553184 CUUUCUCUCAGUGUU--UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU-CACAGCUGCUC ..........(((..--(((((.((.(..((((((((..((....))..))))))))..).)).))((........)))))..)))..-........... ( -29.40, z-score = -0.72, R) >droSec1.super_19 990372 97 - 1257711 CUUUCUCUCAGUGUU--UGCAAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU-CACAGCUGCUC ...............--.(((..((((((((..((((((........((((((((......))).)))))....))))))...)))).-))))..))).. ( -29.10, z-score = -0.98, R) >droEre2.scaffold_4784 18844460 96 - 25762168 CUUUUUCUCAGUGCC--UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCUCAGUCGGAGAAU-AACCGCAUCCACAU-CACAGCUGCUC ............((.--.((..(((.(((((..(((((((((.....((((((((......))).))))))))-))))))...)))))-))).)).)).. ( -32.40, z-score = -1.74, R) >droAna3.scaffold_13337 4878046 94 + 23293914 -UUUCUUCCAGUGUA--UAAAUGAGUGUGUGUGGCGGUUGUUAAAACUUCCGCUGCAGCCUCUGUUGGAGAACAAACCA---CCGCAUCCACAGCUGCUC -.......((((...--.......(((..(((((.(((((((.....(((((..((((...)))))))))))).)))).---)))))..))).))))... ( -24.40, z-score = -0.56, R) >dp4.chrXR_group8 7101766 80 - 9212921 CUGCAUCUGAGUAUCUGUGUGUGUGUGUGUGUGGCGGUAGUUAAAACUUCCGGUGCAUCC--------------AA------CCGCAUCCACAGCUGCUC ........(((((.(((((..((((((..((((.(((.(((....))).))).))))..)--------------).------.))))..))))).))))) ( -30.10, z-score = -3.19, R) >consensus CUUUCUCUCAGUGUU__UGCGAGUGUGUGUGCGGCGGUUGUUAAAACUUCCGCUGCAGCCCCGGUCGGAGAAUAAACCGCAUCCACAU_CACAGCUGCUC ........((((((....))..(((.(((((((((((..((....))..))))))).........(((........))).....)))).))).))))... (-18.64 = -18.43 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:54 2011