| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,012,377 – 19,012,478 |
| Length | 101 |
| Max. P | 0.970678 |

| Location | 19,012,377 – 19,012,478 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Shannon entropy | 0.48821 |
| G+C content | 0.41039 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -12.88 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
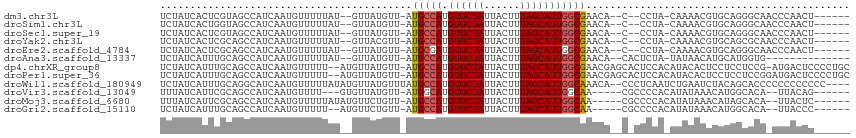
>dm3.chr3L 19012377 101 + 24543557 UCUAUCACUCGUAGCCAUCAAUGUUUUUAU--GUUAUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACA--C--CCUA-CAAAACGUGCAGGGCAACCCAACU------ ..........((.(((....((((((...(--((..((((-.(((((.((((((......)))))))))))))))--.--...)-))))))))....))).))......------ ( -27.00, z-score = -2.57, R) >droSim1.chr3L 18321487 101 + 22553184 UCUAUCACUGGUAGCCAUCAAUGUUUUUAU--GUUAUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACA--C--CCUA-CAAAACGUGCAGGGCAACCCAACU------ .........(((.(((....((((((...(--((..((((-.(((((.((((((......)))))))))))))))--.--...)-))))))))....))).))).....------ ( -30.00, z-score = -2.80, R) >droSec1.super_19 976834 101 + 1257711 UCUAUCACUCGUAGCCAUCAAUGUUUUUAU--GUUAUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACA--C--CCUA-CAAAACGUGCAGGGCAACCCAACU------ ..........((.(((....((((((...(--((..((((-.(((((.((((((......)))))))))))))))--.--...)-))))))))....))).))......------ ( -27.00, z-score = -2.57, R) >droYak2.chr3L 2613485 101 - 24197627 UCUAUCACUCGCAGCCAUCAAUGUUUUUAU--GUUACGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACA--C--CCUA-CAAAACGUGCAGCGCAACCCAACU------ ..........((.((.....((((((...(--((...(((-.(((((.((((((......)))))))))))))).--.--...)-))))))))...)))).........------ ( -21.70, z-score = -1.37, R) >droEre2.scaffold_4784 18830866 101 + 25762168 UCUAUCACUCGCAGCCAUCAAUGUUUUUAU--GUUAUGUU-AUGCGAUGUGCUAUUACUUUAGCAUUGGCGAACA--C--CCUA-CAAAACGUGCAGGGCAACCCAACU------ .............(((....((((((...(--((..((((-.(((.(.((((((......))))))).)))))))--.--...)-))))))))....))).........------ ( -22.30, z-score = -0.67, R) >droAna3.scaffold_13337 4864372 95 - 23293914 UCUAUCAUUUGCAGCCAUCAAUGUUUUUAU--GUUAUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACA--CACUCUA-UAUAACAUGCAUGGUG-------------- .............(((((..(((((..(((--(...((((-.(((((.((((((......)))))))))))))))--.....))-)).)))))..))))).-------------- ( -24.80, z-score = -2.09, R) >dp4.chrXR_group8 7087594 111 + 9212921 UCUAUCAUUUGCAGCCAUCAAUGUUUUU--AUGUUAUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACGAGCACUCCACAUACACUCCUCCUCCG-AUGACUCCCCUGC ..........((((......((((....--.((((.((((-.(((((.((((((......)))))))))))))))))))....))))....((.((....)-).)).....)))) ( -22.10, z-score = -1.67, R) >droPer1.super_36 599848 112 - 818889 UCUAUCAUUUGCAGCCAUCAAUGUUUUU--AUGUUAUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACGAGCACUCCACAUACACUCCUCCUCCGGAUGACUCCCCUGC ..........((((......((((....--.((((.((((-.(((((.((((((......)))))))))))))))))))....)))).((.(((......)))))......)))) ( -25.70, z-score = -2.20, R) >droWil1.scaffold_180949 3077142 109 - 6375548 UCUAUCAUUUGCAGGCAUCAAUGUUUUUAUAUGUUAUGUUUAUGCCAUGUGCUAUUACUUUAGCAUUGGCAAACA--CCCUCAAUCUGAAUCUACAGCACCCCCCCCCCCC---- .........(((((((((.(((((......))))))))))).(((((.((((((......)))))))))))....--...................)))............---- ( -20.90, z-score = -1.88, R) >droVir3.scaffold_13049 11816567 98 - 25233164 UUUAUCAUUCGCAGCCAUCAAUGUUUU---GUGUUAUGUU-AUGGCAUGUGCUAUUACUUUAGCAUUGGCAA-----CGCCCCACAUAUAAACAUGGCACA--UUACAG------ .....................(((..(---((((((((((-(((((..((((((......)))))).(....-----))))......)).)))))))))))--..))).------ ( -25.40, z-score = -1.75, R) >droMoj3.scaffold_6680 13928555 101 + 24764193 UUUAUCAUUCGCAGCCAUCAAUGUUUUUAUAUGUUCUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCAA-----CGCCCCACAUAUAAACAUAGCACA--UUACUC------ .....................(((.(((((((((...((.-.(((((.((((((......))))))))))).-----.))...))))))))).))).....--......------ ( -22.30, z-score = -2.82, R) >droGri2.scaffold_15110 5713289 99 - 24565398 UCUAUCAUUUGCAGCCAUCAAUGUUUUU--AUGUUCUGUU-AUGCCAUGUGCUAUUACUUUAGCAUUGGCAA-----CGCCCCACAUAUAAACAUGGCACA--UUACCC------ .............((((....(((((.(--((((...((.-.(((((.((((((......))))))))))).-----.))...))))).)))))))))...--......------ ( -27.50, z-score = -3.65, R) >consensus UCUAUCAUUCGCAGCCAUCAAUGUUUUUAU__GUUAUGUU_AUGCCAUGUGCUAUUACUUUAGCAUUGGCGAACA__C_CCCUA_CAAAACGUGCAGCGCA_CCCACCC______ ..........................................(((((.((((((......)))))))))))............................................ (-12.88 = -12.82 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:51 2011