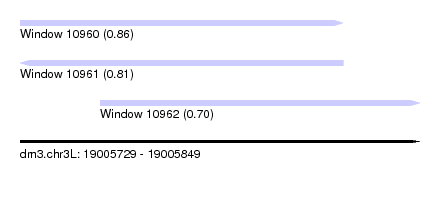
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 19,005,729 – 19,005,849 |
| Length | 120 |
| Max. P | 0.862902 |

| Location | 19,005,729 – 19,005,826 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.55 |
| Shannon entropy | 0.51837 |
| G+C content | 0.46148 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -15.61 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.862902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19005729 97 + 24543557 -----GCCAAGGACCCCGCCCAUUUG-----------AGUAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUA- -----.....((.......)).....-----------.(((((.(...((((...(((((...((((((((...))))))))..))))).))))...))))))..........- ( -23.80, z-score = -1.61, R) >droSim1.chr3L 18314923 97 + 22553184 -----GCCAAGGACCCCGCCCAUUUA-----------UGCAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUA- -----.....((.......))...((-----------(((((((....((((...(((((...((((((((...))))))))..))))).))))...(....).)))))))))- ( -22.60, z-score = -1.04, R) >droSec1.super_19 970307 97 + 1257711 -----GCCAAGGACCCCGCCCAUUUG-----------AGCAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUA- -----.....((((...((.(....)-----------.))........((((...(((((...((((((((...))))))))..))))).))))...))))............- ( -22.60, z-score = -0.99, R) >droYak2.chr3L 2601792 96 - 24197627 -----GCCAAGGACCCCGCCCAUUUG-----------AGUAGAG-CCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUA- -----.....((.......)).....-----------.(((((.-...((((...(((((...((((((((...))))))))..))))).))))....)))))..........- ( -24.40, z-score = -1.79, R) >dp4.chrXR_group8 7080713 88 + 9212921 -----------------GCCGGGCAGG-------CCCCGCCCAG--CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUUUCCAUUUUGUGUAA -----------------((.((((.(.-------...))))).)--).((((...(((((...((((((((...))))))))..))))).)))).................... ( -28.30, z-score = -1.56, R) >droPer1.super_36 592965 88 - 818889 -----------------GCCGGGCAGG-------CCCCGCCCAG--CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUUUCCAUUUUGUGUAA -----------------((.((((.(.-------...))))).)--).((((...(((((...((((((((...))))))))..))))).)))).................... ( -28.30, z-score = -1.56, R) >droAna3.scaffold_13337 4858098 108 - 23293914 -----GCCAAGGACCCCGCCCAUUUUCAGUGUUUUCCAGCUCAGCCGAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUCCGUUUUGUAUA- -----.....((((..............((((((((..((...)).)))))))).(((((...((((((((...))))))))..))))).))))...................- ( -23.90, z-score = -0.70, R) >droEre2.scaffold_4784 18824176 97 + 25762168 -----GCCAAGGACCCCGCCCAUUUG-----------AGUAGAGACCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCACAGCAGUCCCAAGUCUACAUUUUGUAUA- -----.....((.......)).....-----------.(((((.....((((...(((.....((((((((...))))))))....))).))))....)))))..........- ( -21.40, z-score = -0.97, R) >droWil1.scaffold_180949 3307501 84 + 6375548 -----------------GCCCAGUUG-----------UGU-GAGGCCAGGACAUGGCUUUUAAUUGCAUGAGCAUUAUGCAAGCAAAGCAUUCCCAAGUUUCCGUUUUGCAGA- -----------------.........-----------...-(((((((.....)))))))...((((((((...))))))))(((((((..............)))))))...- ( -21.34, z-score = -0.10, R) >droMoj3.scaffold_6680 13919626 106 + 24764193 GCUAAGCUGACCCAAUUUCUCAUUUUUU---UUUUUCGCUCUGCACCAGGACACGCUUUUUAAUUGCAUAAGCAUUAUGUAA--AAGGAGUCCCCAGCUAGUUGCUUUGUC--- ((((.((((...................---......((...))....((((...(((((((..(((....))).....)))--)))).)))).)))))))).........--- ( -20.00, z-score = -0.26, R) >consensus _____GCCAAGGACCCCGCCCAUUUG___________AGCAGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUA_ ................................................((((...(((((...((((((((...))))))))..))))).)))).................... (-15.61 = -15.75 + 0.14)

| Location | 19,005,729 – 19,005,826 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.55 |
| Shannon entropy | 0.51837 |
| G+C content | 0.46148 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -14.54 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.806354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19005729 97 - 24543557 -UAUACAAAAUGUAGACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGGCUCUACU-----------CAAAUGGGCGGGGUCCUUGGC----- -....(((...........(((((.(((((..((((((.......))))))...)))))...)))))((((((..((-----------(....)))..)))))))))..----- ( -31.20, z-score = -2.29, R) >droSim1.chr3L 18314923 97 - 22553184 -UAUACAAAAUGUAGACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGGCUCUGCA-----------UAAAUGGGCGGGGUCCUUGGC----- -....(((.(((((((((..((((.(((((..((((((.......))))))...)))))...))))..)).))))))-----------)....((((...)))))))..----- ( -33.70, z-score = -2.70, R) >droSec1.super_19 970307 97 - 1257711 -UAUACAAAAUGUAGACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGGCUCUGCU-----------CAAAUGGGCGGGGUCCUUGGC----- -....(((...........(((((.(((((..((((((.......))))))...)))))...)))))((((((((((-----------(....))))))))))))))..----- ( -36.50, z-score = -3.67, R) >droYak2.chr3L 2601792 96 + 24197627 -UAUACAAAAUGUAGACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGG-CUCUACU-----------CAAAUGGGCGGGGUCCUUGGC----- -....(((...(((((.(..((((.(((((..((((((.......))))))...)))))...))))..)-.))))).-----------.....((((...)))))))..----- ( -29.00, z-score = -1.82, R) >dp4.chrXR_group8 7080713 88 - 9212921 UUACACAAAAUGGAAACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUG--CUGGGCGGGG-------CCUGCCCGGC----------------- ...........(....)..(((((.(((((..((((((.......))))))...)))))...)))))(--((((((((..-------.)))))))))----------------- ( -32.80, z-score = -2.49, R) >droPer1.super_36 592965 88 + 818889 UUACACAAAAUGGAAACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUG--CUGGGCGGGG-------CCUGCCCGGC----------------- ...........(....)..(((((.(((((..((((((.......))))))...)))))...)))))(--((((((((..-------.)))))))))----------------- ( -32.80, z-score = -2.49, R) >droAna3.scaffold_13337 4858098 108 + 23293914 -UAUACAAAACGGAGACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUCGGCUGAGCUGGAAAACACUGAAAAUGGGCGGGGUCCUUGGC----- -..........(((..((((((((.(((((..((((((.......))))))...)))))...))))(((((...))))).................)))).))).....----- ( -25.40, z-score = 0.40, R) >droEre2.scaffold_4784 18824176 97 - 25762168 -UAUACAAAAUGUAGACUUGGGACUGCUGUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGUCUCUACU-----------CAAAUGGGCGGGGUCCUUGGC----- -....(((...........(((((.(((....((((((.......)))))).....)))...)))))((.(((..((-----------(....)))..))).)))))..----- ( -25.50, z-score = -0.53, R) >droWil1.scaffold_180949 3307501 84 - 6375548 -UCUGCAAAACGGAAACUUGGGAAUGCUUUGCUUGCAUAAUGCUCAUGCAAUUAAAAGCCAUGUCCUGGCCUC-ACA-----------CAACUGGGC----------------- -((((.....)))).....((((..(((((..((((((.......))))))...)))))....)))).(((.(-(..-----------....)))))----------------- ( -19.10, z-score = -0.06, R) >droMoj3.scaffold_6680 13919626 106 - 24764193 ---GACAAAGCAACUAGCUGGGGACUCCUU--UUACAUAAUGCUUAUGCAAUUAAAAAGCGUGUCCUGGUGCAGAGCGAAAAA---AAAAAAUGAGAAAUUGGGUCAGCUUAGC ---....((((.....(((.(((((.((((--((...((((((....)).))))))))).).))))).).)).((.(.((...---.............)).).)).))))... ( -20.49, z-score = 0.71, R) >consensus _UAUACAAAAUGUAGACUUGGGACUGCUUUGCUUGCAUAAUGUUCAUGCAAUUAAAAGCCAUGUCCUGGGCUCUACU___________CAAAUGGGCGGGGUCCUUGGC_____ ...................(((((.(((((..((((((.......))))))...)))))...)))))............................................... (-14.54 = -14.87 + 0.33)


| Location | 19,005,753 – 19,005,849 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Shannon entropy | 0.39548 |
| G+C content | 0.41858 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -16.93 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.701965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 19005753 96 + 24543557 AGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUGUUGGUUGGUCGCCUUCCC-------- .((.((((((((...(((((...((((((((...))))))))..))))).)))).((((.(((.....))).))))...)))..).))........-------- ( -24.60, z-score = -1.05, R) >droSim1.chr3L 18314947 96 + 22553184 AGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUGUUGGUUGGUCGCCUUCCC-------- .((.((((((((...(((((...((((((((...))))))))..))))).)))).((((.(((.....))).))))...)))..).))........-------- ( -24.60, z-score = -1.05, R) >droSec1.super_19 970331 96 + 1257711 AGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUGUUGGUUGGUCGCCUUCCC-------- .((.((((((((...(((((...((((((((...))))))))..))))).)))).((((.(((.....))).))))...)))..).))........-------- ( -24.60, z-score = -1.05, R) >droYak2.chr3L 2601816 95 - 24197627 AGAG-CCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUGUUGGUUGGUCGCCUUCCC-------- ..((-(((((((...(((((...((((((((...))))))))..))))).)))).((((.(((.....))).))))...)))))............-------- ( -25.80, z-score = -1.47, R) >dp4.chrXR_group8 7080733 97 + 9212921 ------CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUUUCCAUUUUGUGUA-AUUUUCCCUUCAUUCUUUCAUUUCUUUCUG ------..((((...(((((...((((((((...))))))))..))))).))))...................-.............................. ( -17.70, z-score = -1.36, R) >droPer1.super_36 592985 97 - 818889 ------CAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUUUCCAUUUUGUGUA-AUUUUCCCUUCAUUCUUUCAUUUCUUUCUG ------..((((...(((((...((((((((...))))))))..))))).))))...................-.............................. ( -17.70, z-score = -1.36, R) >droAna3.scaffold_13337 4858133 103 - 23293914 UCAGCCGAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUCCGUUUUGUAUACUUUGUUGGUCGCCUUUGUAGGUAAUGCCA- ...(((((((((...(((((...((((((((...))))))))..))))).))))((((.......)))).........))))).((((....)))).......- ( -25.30, z-score = -0.42, R) >droEre2.scaffold_4784 18824200 96 + 25762168 AGAGACCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCACAGCAGUCCCAAGUCUACAUUUUGUAUACUUUGUUGGUUGGUCGCCUUCCC-------- ...(((((((((...(((.....((((((((...))))))))....))).)))).((((.(((.....))).)))).......)))))........-------- ( -26.20, z-score = -1.67, R) >droWil1.scaffold_180949 3307512 96 + 6375548 UGAGGCCAGGACAUGGCUUUUAAUUGCAUGAGCAUUAUGCAAGCAAAGCAUUCCCAAGUUUCCGUUUUGCAGACAUUUUCCCCCCAUUUCUUUUUC-------- .(((((((.....)))))))...((((((((...))))))))(((((((..............)))))))..........................-------- ( -21.34, z-score = -1.19, R) >consensus AGAGCCCAGGACAUGGCUUUUAAUUGCAUGAACAUUAUGCAAGCAAAGCAGUCCCAAGUCUACAUUUUGUAUACUUUGUUGGUUGGUCGCCUUCCC________ ........((((...(((((...((((((((...))))))))..))))).)))).................................................. (-16.93 = -17.16 + 0.22)

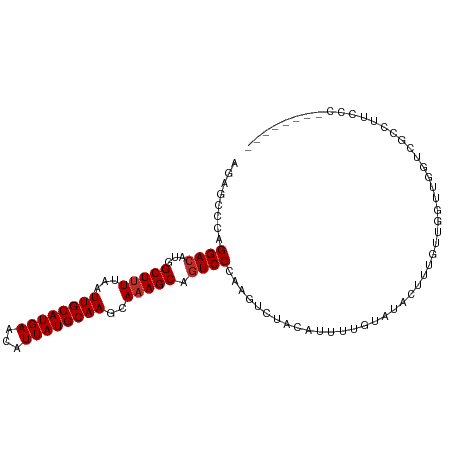
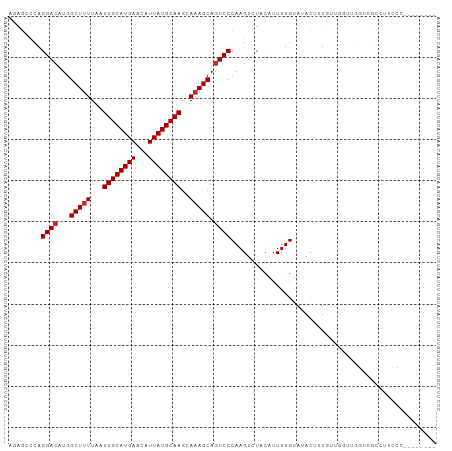
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:49 2011