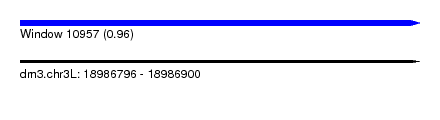
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,986,796 – 18,986,900 |
| Length | 104 |
| Max. P | 0.962158 |

| Location | 18,986,796 – 18,986,900 |
|---|---|
| Length | 104 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 59.67 |
| Shannon entropy | 0.80223 |
| G+C content | 0.49687 |
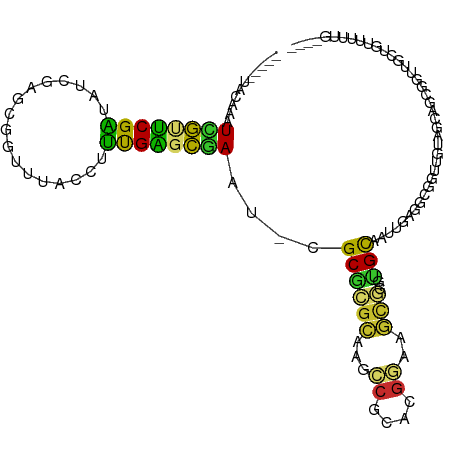
| Mean single sequence MFE | -35.59 |
| Consensus MFE | -12.95 |
| Energy contribution | -11.35 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.962158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
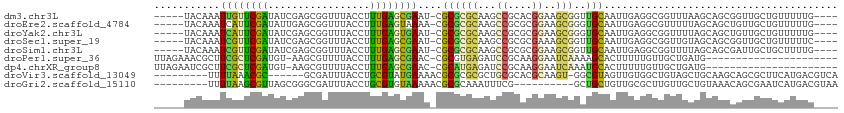
>dm3.chr3L 18986796 104 + 24543557 -----UACAAAUUGUUCGAUAUCGAGCGGUUUACCUUUGAGCGAAU-CGCGCGCAAGCCGCACGGAAGCGGUUGCAAUUGAGGCGGUUUAAGCAGCGGUUGCUGUUUUUG---- -----...((((((((((....)))))))))).......(((.(((-(((((..(((((((.(((..((....))..)))..)))))))..)).))))))))).......---- ( -35.80, z-score = -0.97, R) >droEre2.scaffold_4784 18805570 104 + 25762168 -----UACAAAUCAUUCGAUAUUGAGCGGUUUACCUUUGAGUAAAA-CGCGCGCAAGCCGCGCGGAAGCGGGUGCAAUUGAGGCGUUUUUAGCAGCUGUUGCUGUUUUUG---- -----..((((...((((((.(((.(((.(((((......))))).-)))...)))(((.(((....))).).)).))))))........((((((....))))))))))---- ( -30.90, z-score = 0.07, R) >droYak2.chr3L 2582949 104 - 24197627 -----UACAAAUCAUUCGAUAUCGAGCGGUUUACCUUUGAGUGAAU-CGCGCGCAAGCCGCGCGGAAGCGGGUGCAAUUGAGGCGGUUUUAGCAGCUGUUGCUGUUUUUG---- -----...(((((.((((((...(.(((((((((......))))))-))).)((..(((.(((....)))))))).))))))..))))).((((((....))))))....---- ( -37.40, z-score = -1.42, R) >droSec1.super_19 951740 104 + 1257711 -----UACAAAUCGUUCGAUAUCGAGCGGUUUACCUUUGAGCGAAU-CGCGCGCAAGCCGCGCGAAAGCGGUUGCAAUUGAGGCGGUUGUAGCAGCGGUUGCUGUUUUUC---- -----...((((((((((....)))))))))).......(((.(((-(((((....))..(((....)))((((((((((...)))))))))).))))))))).......---- ( -42.80, z-score = -2.35, R) >droSim1.chr3L 18295718 104 + 22553184 -----UACAAAUCGUUCGAUAUCGAGCGGUUUACCUUUGAGCGAAU-CGCGCGCAAGCCGCGCGGAAGCGGUUGCAAUUGAGGCGGUUUUAGCAGCGAUUGCUGCUUUUG---- -----...((((((((((....))))))))))......((((((((-(((((....).)))((....))))))(((((((..((.......))..))))))))))))...---- ( -41.40, z-score = -1.71, R) >droPer1.super_36 574467 90 - 818889 UUAGAAACGCUUCGCUCGAUGU-AAGCGUUUUACCUUUGAGCGAAC-CGCGUGAGAUCCGCAAGGAAUCAAAAGCACUUUUUGUUGCUGAUG---------------------- ((((.((((.(((((((((.((-(((...)))))..))))))))).-)((.(((..(((....))).)))...)).......))).))))..---------------------- ( -29.00, z-score = -2.62, R) >dp4.chrXR_group8 7062336 90 + 9212921 UUAGAAUCGCUUCGCUCGAUGU-AAGCGUUUUACCUUUGAGCGAAC-CGCAUGAGAUCCGCAAGGAAUCAAAUGCACUUUUUGUUGCUGAUG---------------------- .....((((((((((((((.((-(((...)))))..))))))))).-.((((..(((((....)).)))..))))..........)).))).---------------------- ( -28.30, z-score = -2.73, R) >droVir3.scaffold_13049 11791050 98 - 25233164 ---------UUUUAAACGC------GCGAUUUACCUGCGUAUGAAAACGCGCGCGCUGCGCACGCAAGU-GGCGUAGUUGUGGCUGUAGCUGCAAGCAGCGCUUCAUGACGUCA ---------........((------((.........(((..(....)..)))(((((((((((....))-.)))))))((..((....))..)).)).))))............ ( -37.20, z-score = 0.51, R) >droGri2.scaffold_15110 5686261 95 - 24565398 ---------UUUUAAGCGUUAGCGGGCGAUUUACCUGCGUGUAAAAACGCGCAAAUUUCG----------GCUGCUGUUGCGCUUGUUGCUGUAAACAGCGAAUCAUGACGUAA ---------...(((((((((((((.(((......(((((((....)))))))....)))----------.))))))..)))))))((((((....))))))............ ( -37.50, z-score = -3.08, R) >consensus _____UACAAAUCGUUCGAUAUCGAGCGGUUUACCUUUGAGCGAAU_CGCGCGCAAGCCGCACGGAAGCGGGUGCAAUUGAGGCGGUUGUAGCAGCGGUUGCUGUUUUUG____ ...........((((((((.................))))))))....((((((...((....))..)))..)))....................................... (-12.95 = -11.35 + -1.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:45 2011