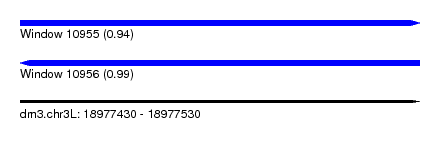
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,977,430 – 18,977,530 |
| Length | 100 |
| Max. P | 0.993609 |

| Location | 18,977,430 – 18,977,530 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Shannon entropy | 0.36602 |
| G+C content | 0.47008 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -14.55 |
| Energy contribution | -14.69 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
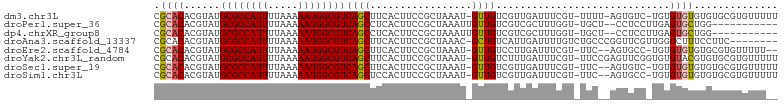
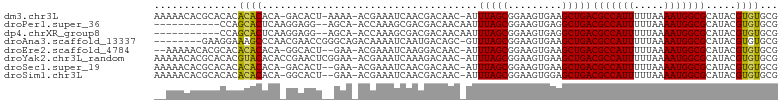
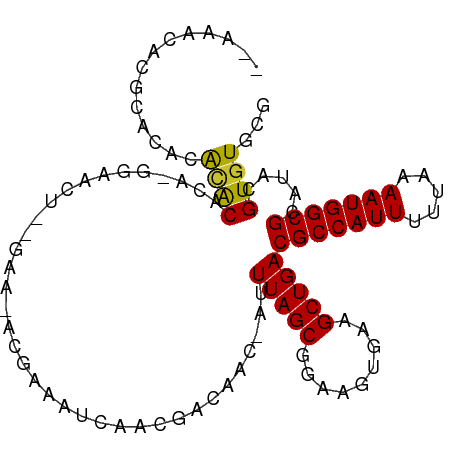
>dm3.chr3L 18977430 100 + 24543557 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUUCACUUCCGCUAAAU-GUUGUCGUUGAUUUCGU-UUUU-AGUGUC-UGUGUGUGUGUGCGUGUUUUU (((((((....((((((((.....))))))))..((..(((...((((((((-(..(((...)))..)))-..))-))))..-.))).)))))))))....... ( -32.10, z-score = -3.63, R) >droPer1.super_36 565087 90 - 818889 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCCUCACUUCCGCUAAAUUGUUGUCGUCGCUUUGGU-UGCU--CCUCCUUGAGUGCUGG----------- .((((......((((((((.....))))))))(((((.......((.(((....))).)).......)))-))..--.........))))...----------- ( -21.44, z-score = -0.29, R) >dp4.chrXR_group8 7053029 90 + 9212921 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCCUCACUUCCGCUAAAUUGUUGUCGUCGCUUUGGU-UGCU--CCUCCUUGAGUGCUGG----------- .((((......((((((((.....))))))))(((((.......((.(((....))).)).......)))-))..--.........))))...----------- ( -21.44, z-score = -0.29, R) >droAna3.scaffold_13337 4830640 95 - 23293914 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUUCACUUCCGCUAAAC-GCUGUCAUUGAUUUUGUCUGCCCGGUUCGUUGGGCUUUCCUUC-------- .(.((((((..((((((((.....)))))))).(((.........)))..))-).)))).............((((((....))))))........-------- ( -22.90, z-score = -0.65, R) >droEre2.scaffold_4784 18796291 97 + 25762168 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUUCACUUCCGCUAAAU-GUUGUCCUUGAUUUCGU-UUC--AGUGCC-UGUGUGUGUGCGUGUUUUU-- (((((((((..((((((((.....))))))))..))..(((...((((((((-(..(((...)))..)))-)).--))))..-.))).))))))).......-- ( -33.00, z-score = -4.29, R) >droYak2.chr3L_random 2862519 102 + 4797643 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUUCACUUCCGCUAAAU-GUUGUCUUUGAUUUCGU-UUCCGAGUUCGGUGUGUACGUGUGCGUGUUUUU ((((((((((((((((..........(((((..((.....))..)))))...-.........((.((((.-...)))).))))))))))))))))))....... ( -38.50, z-score = -5.09, R) >droSec1.super_19 942492 99 + 1257711 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUUCACUUCCGCUAAAU-GUUGUCGUUGAUUUCGU-UUC--AGUGUC-UGUGUGUGUGUGCGUGUUUUU (((((((....((((((((.....))))))))..((..(((...((((((((-(..(((...)))..)))-)).--))))..-.))).)))))))))....... ( -33.00, z-score = -3.74, R) >droSim1.chr3L 18286348 99 + 22553184 CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUCCACUUCCGCUAAAU-GUUGUCGUUGAUUUCGU-UUC--AGUGCC-UGUGUGUGUGUGCGUGUUUUU (((((((((..((((((((.....))))))))..))..(((...((((((((-(..(((...)))..)))-)).--))))..-.)))...)))))))....... ( -33.20, z-score = -3.61, R) >consensus CGCACACGUAUGCGCCAUUUUAAAAAUGGCGUCAGCUUCACUUCCGCUAAAU_GUUGUCGUUGAUUUCGU_UUC__AGUGCC_UGUGUGUGUGUGCGUGUUU__ .((((......((((((((.....)))))))).(((.........)))......................................)))).............. (-14.55 = -14.69 + 0.14)
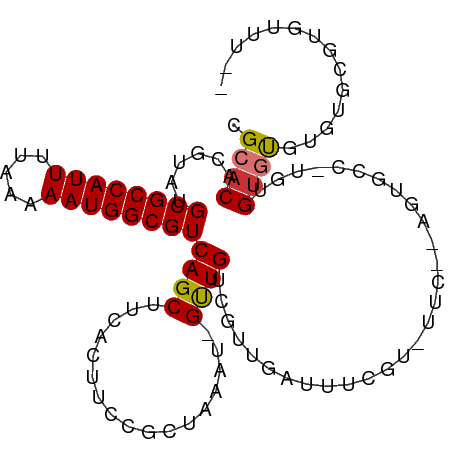
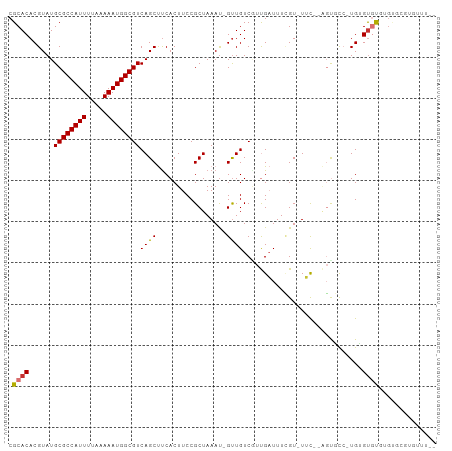
| Location | 18,977,430 – 18,977,530 |
|---|---|
| Length | 100 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.01 |
| Shannon entropy | 0.36602 |
| G+C content | 0.47008 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -15.92 |
| Energy contribution | -15.61 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18977430 100 - 24543557 AAAAACACGCACACACACACA-GACACU-AAAA-ACGAAAUCAACGACAAC-AUUUAGCGGAAGUGAAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG .......(((((((...(((.-..(.((-(((.-.................-.))))).)...)))....((.(((((((.....)))))))))...))))))) ( -25.81, z-score = -3.63, R) >droPer1.super_36 565087 90 + 818889 -----------CCAGCACUCAAGGAGG--AGCA-ACCAAAGCGACGACAACAAUUUAGCGGAAGUGAGGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG -----------.((((.(((..(..(.--.((.-......))..)..).........((....)))))))))..((((((.....))))))((((....)))). ( -28.30, z-score = -2.49, R) >dp4.chrXR_group8 7053029 90 - 9212921 -----------CCAGCACUCAAGGAGG--AGCA-ACCAAAGCGACGACAACAAUUUAGCGGAAGUGAGGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG -----------.((((.(((..(..(.--.((.-......))..)..).........((....)))))))))..((((((.....))))))((((....)))). ( -28.30, z-score = -2.49, R) >droAna3.scaffold_13337 4830640 95 + 23293914 --------GAAGGAAAGCCCAACGAACCGGGCAGACAAAAUCAAUGACAGC-GUUUAGCGGAAGUGAAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG --------........((((........))))...............((((-.....((....))...))))..((((((.....))))))((((....)))). ( -28.60, z-score = -1.97, R) >droEre2.scaffold_4784 18796291 97 - 25762168 --AAAAACACGCACACACACA-GGCACU--GAA-ACGAAAUCAAGGACAAC-AUUUAGCGGAAGUGAAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG --.......(((((((...((-(.((((--...-.((.......(.....)-......))..))))...))).(((((((.....))))))).....))))))) ( -27.22, z-score = -3.03, R) >droYak2.chr3L_random 2862519 102 - 4797643 AAAAACACGCACACGUACACACCGAACUCGGAA-ACGAAAUCAAAGACAAC-AUUUAGCGGAAGUGAAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG .......((((((((((......((..(((...-.)))..)).........-..(((((.........)))))(((((((.....)))))))..)))))))))) ( -35.10, z-score = -5.36, R) >droSec1.super_19 942492 99 - 1257711 AAAAACACGCACACACACACA-GACACU--GAA-ACGAAAUCAACGACAAC-AUUUAGCGGAAGUGAAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG .......(((((((.....((-(.((((--...-.((..............-......))..))))...))).(((((((.....))))))).....))))))) ( -26.65, z-score = -3.47, R) >droSim1.chr3L 18286348 99 - 22553184 AAAAACACGCACACACACACA-GGCACU--GAA-ACGAAAUCAACGACAAC-AUUUAGCGGAAGUGGAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG .......(((((((.......-(((.((--...-.((.......)).....-.....((....)))).)))(.(((((((.....))))))))....))))))) ( -26.80, z-score = -2.69, R) >consensus __AAACACGCACACACACACA_GGAACU__GAA_ACGAAAUCAACGACAAC_AUUUAGCGGAAGUGAAGCUGACGCCAUUUUUAAAAUGGCGCAUACGUGUGCG ..............((((....................................(((((.........)))))(((((((.....))))))).....))))... (-15.92 = -15.61 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:44 2011