
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,970,407 – 18,970,481 |
| Length | 74 |
| Max. P | 0.888118 |

| Location | 18,970,407 – 18,970,481 |
|---|---|
| Length | 74 |
| Sequences | 8 |
| Columns | 85 |
| Reading direction | forward |
| Mean pairwise identity | 60.08 |
| Shannon entropy | 0.74221 |
| G+C content | 0.52211 |
| Mean single sequence MFE | -22.71 |
| Consensus MFE | -10.14 |
| Energy contribution | -9.38 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.86 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.888118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18970407 74 + 24543557 CGAGUCCAGGAGCUUGCAUAGAUUUGCGACGUUUUUCACGUCCCUCGAGGGACGCGAGACGCGAACUGGGUGUG----------- ((..(((((....(((((......)))))(((.((((.((((((....)))))).)))).)))..)))))..))----------- ( -29.90, z-score = -2.47, R) >droSim1.chr3L 18276598 74 + 22553184 CGAGUCCAGGAGCUUGCAUAGAUUUGCGACGUUUUUCACGUCCCUCGAGGGACGCGGGACGCGAAGUGGGUGUG----------- ((.(((((((((((((((......))))).))))))(.((((((....)))))).))))).))...........----------- ( -27.70, z-score = -1.45, R) >droSec1.super_19 935730 74 + 1257711 CGAGUCCAGGAGCUUGCAUAGAUUUGCGACGUUUUUCACGUCCCUCGAGGGACGCGGGACGCGAUGUGGGUGUG----------- ((.(((((((((((((((......))))).))))))(.((((((....)))))).))))).))...........----------- ( -27.70, z-score = -1.39, R) >droYak2.chr3L_random 2844600 74 + 4797643 CGAGUCCAGGAGCUUGCAUAGAUUUGCGACGUUUUUCAUGUUCCCCAAGGGAUGCGGGACGGAUAAUGGGUGUG----------- ...(((((((((((((((......))))).))))))..(((((((...)))).)))))))..............----------- ( -19.10, z-score = 0.24, R) >droEre2.scaffold_4784 18789518 74 + 25762168 CGAGUCCAGGAGCUUGCAUAGAUUUGCGACGUUUUUCAUGUUCCCCAAGGGAUGCGGGAGGAGAAGUAGAUGUG----------- (((((......)))))..((.((((((....((((((.(((((((...)))).))).))))))..)))))).))----------- ( -17.00, z-score = 0.30, R) >dp4.chrXR_group8 7045152 79 + 9212921 -----GGAGCUCCUUGCAUAGAUUUGCGACGUCUCCCUUGGGG-GAAAUUCCAAAGGGAAAAACAGUGAGAGAGCAAGAGGGGUG -----...((((((((((......)))......((((((((((-....)))).))))))..................))))))). ( -23.40, z-score = -0.63, R) >droPer1.super_36 557244 79 - 818889 -----GGAGCUCCUUGCAUAGAUUUGCGACGUCUCCCUUGGGG-GAAAUUCCAAAGGGAAAAACAGUGAGAGAGCAAGAGGGGUG -----...((((((((((......)))......((((((((((-....)))).))))))..................))))))). ( -23.40, z-score = -0.63, R) >droMoj3.scaffold_6680 13881875 64 + 24764193 AAAGCCAAAGAGCUUUCAUAGAGUUU---CAUUUCCUUUUGGU-UAAAUGAAACCUGACUAGUUUCUA----------------- (((((......)))))..(((.((((---(((((((....)).-.))))))))))))...........----------------- ( -13.50, z-score = -1.74, R) >consensus CGAGUCCAGGAGCUUGCAUAGAUUUGCGACGUUUUUCAUGUCCCUCAAGGGAAGCGGGACGAGAAGUGGGUGUG___________ .............(((((......)))))....((((.(((((......))))).)))).......................... (-10.14 = -9.38 + -0.77)

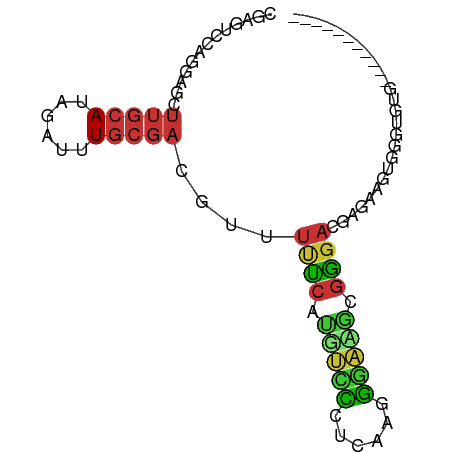
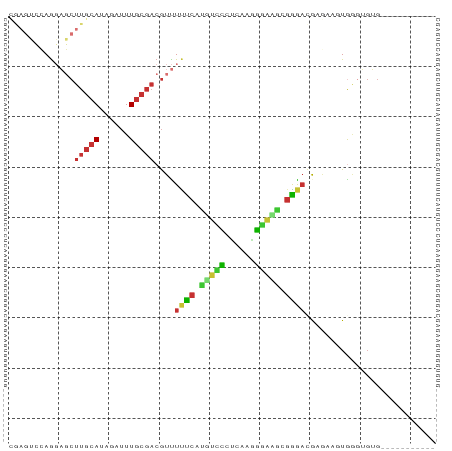
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:43 2011