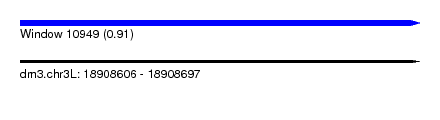
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,908,606 – 18,908,697 |
| Length | 91 |
| Max. P | 0.911518 |

| Location | 18,908,606 – 18,908,697 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 66.88 |
| Shannon entropy | 0.67625 |
| G+C content | 0.46561 |
| Mean single sequence MFE | -15.56 |
| Consensus MFE | -8.21 |
| Energy contribution | -8.06 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.911518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
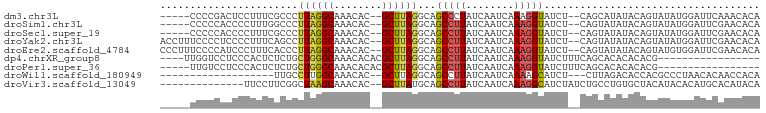
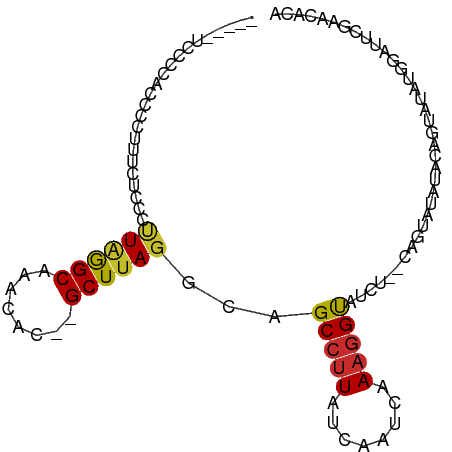
>dm3.chr3L 18908606 91 + 24543557 -----CCCCGACUCCUUUCGCCCUUAGGCAAACAC--GCUUAGGCAGCCCUAUCAAUCAAAGGUAUCU--CAGCAUAUACAGUAUAUGGAUUCAAACACA -----....((.(((....(((....)))......--(((.(((..(((............))).)))--.))).............))).))....... ( -15.00, z-score = -0.50, R) >droSim1.chr3L 18214589 91 + 22553184 -----CCCCCACCCCUUUGGCCCUUAGGCAAACAC--GCUUAGGCAGCCUUAUCAAUCAAAGGUAUCU--CAGUAUAUACAGUAUAUGGAUUCGAACACA -----...((...((((((((.(((((((......--)))))))..))).........))))).....--..((((((...))))))))........... ( -16.10, z-score = 0.15, R) >droSec1.super_19 874303 91 + 1257711 -----CCCCCACCCCUUUCGCCCUUAGGCAAACAC--GCUUAGGCAGCCUUAUCAAUCAAAGGUAUCU--CAGUAUAUACAGUAUAUGGAUUCGAACACA -----...(((........(((..(((((......--)))))))).(((((........)))))....--................)))........... ( -15.60, z-score = -0.68, R) >droYak2.chr3L 2487208 96 - 24197627 ACCUUUCCCCUCCCCUUUCAGCCUUAGGCAAACAC--GCUUAGGCAGCCUUAUCAAUCAAAGGUAUCU--CAGUAUAUACAGUAUAUGGAUUCGAACACA ....(((...(((.......((((.((((......--)))))))).(((((........)))))....--..((((((...)))))))))...))).... ( -17.70, z-score = -1.27, R) >droEre2.scaffold_4784 18726253 96 + 25762168 CCCUUUCCCCAUCCCUUUCACCCUUAGGCAAACAC--GCUUAGGCAGCCUUAUCAAUCAAAGGUAUCU--CAGUAUAUACAGUAUGUGGAUUCGAACACA ...........((...(((((.(((((((......--)))))))..(((((........)))))....--...............)))))...))..... ( -15.40, z-score = -0.40, R) >dp4.chrXR_group8 6981173 79 + 9212921 ----UUGGUCCUCCCACUCUCUGCUGGGCAAACACACGCUUAGGCAGCCUUAUCAAUCAAAGGUAUCUUUCAGCACACACACG----------------- ----.(((.....))).....(((((((.........((....)).(((((........)))))....)))))))........----------------- ( -14.20, z-score = -0.56, R) >droPer1.super_36 493033 78 - 818889 -----UUGUCCUCCCACUCUCUGCUGGGCAAACACACGCUUAGGCAGCCUUAUCAAUCAAAGGUAUCUUUCAGCACACACACG----------------- -----((((((..............))))))......(((.(((..(((((........)))))..)))..))).........----------------- ( -13.94, z-score = -0.99, R) >droWil1.scaffold_180949 3430570 76 + 6375548 -------------------UUGCCUUGGCAAACAC--GCUUAGGCAGCCUUAUCAAUCAAAAGCAUCU---CUUAGACACCACGCCCUAACACAACCACA -------------------((((((.(((......--))).)))))).....................---............................. ( -11.40, z-score = -1.01, R) >droVir3.scaffold_13049 11698331 84 - 25233164 --------------UUCCUUCGGCUAAGCAAACAC--GCUUAUGCAGCCUUAUCAAUCAAAGGCAUCUAUCUGCCUGUGCUACAUACACAUGCACAUACA --------------.......((((((((......--)))))....(((((........)))))........)))(((((...........))))).... ( -20.70, z-score = -2.93, R) >consensus _____UCCCCACCCCUUUCUCCCUUAGGCAAACAC__GCUUAGGCAGCCUUAUCAAUCAAAGGUAUCU__CAGUAUAUACAGUAUAUGGAUUCGAACACA .......................((((((........))))))...(((((........))))).................................... ( -8.21 = -8.06 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:39 2011