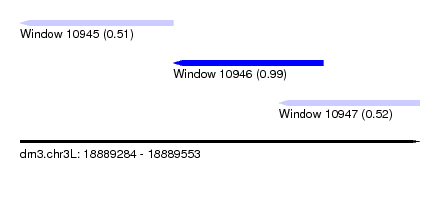
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,889,284 – 18,889,553 |
| Length | 269 |
| Max. P | 0.994041 |

| Location | 18,889,284 – 18,889,387 |
|---|---|
| Length | 103 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 58.34 |
| Shannon entropy | 0.89061 |
| G+C content | 0.45122 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -4.64 |
| Energy contribution | -3.77 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.69 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.16 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.509462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18889284 103 - 24543557 UCCCUAGAGAGUUGAAUUACCUGAGUGGGGGGAGGACGACGACUUGUUGUUGUUGUCCCCCUGCAAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUUUCCUU- ..(((((..((..((((((..((.(..((((((.((((((((....)))))))).))))))..)....))..))))))))..)))))((((........))))- ( -39.90, z-score = -3.13, R) >droSim1.chr3L 18194827 103 - 22553184 UCCCUAGAGAGUUGAAUUACCUGAGUGGGGGGAGAAGAACGACUUGUUGUUGUUCUCCCCCUGCAAGCCAUCUAAUUCCUAGUUGGGAAGGUUACAUUUCCUU- ...((((.((((((.((...((..(..(((((((((.(((((....))))).)))))))))..).))..)).))))))))))..((((((......)))))).- ( -41.00, z-score = -3.66, R) >droSec1.super_19 854905 103 - 1257711 UCCCUAGAGAGUUGAAUUACCUGAGUGGGCGGAGAAGAACGACUUGUUGUUGUUCUCCCCCUGCAAGCCAUCUAAUUCCUAGUUGAGAAGGUUACAUUUCCUU- ...((((.((((((.((..(.((...(((.((((((.(((((....))))).)))))))))..)).)..)).)))))))))).....((((........))))- ( -31.60, z-score = -2.51, R) >droYak2.chr3L 2447920 85 + 24197627 UCCCUAGAGAGUUGAAUUACCUGAGCGGGAG---------GACUCG---------UCCUCCUGCCAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUUUCUUU- .....((((((.((....(((((.(((((((---------((....---------))))))))))..(((.(((.....))).)))..))))..)).))))))- ( -29.60, z-score = -2.52, R) >droEre2.scaffold_4784 18705708 79 - 25762168 UCCCUAGAGAGUUGAAUUACCU----GGGAG---------GACUCG---------UCCUCCUGCCAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUCCUU--- (((((((.(((((((.......----(((((---------((....---------)))))))........)).))))))))...))))(((......))).--- ( -22.56, z-score = -0.53, R) >droAna3.scaffold_13337 4742154 84 + 23293914 UCCCUAGAGAGUUGAGCCAAUUGAGUGGGGGUG-------UAC------------UUCUCCUGCAAUCCAACUGGCUCCUAGUUGGGAUGGUUACAUUUUCUU- .....(((((((..(((((.....(..((((..-------...------------..))))..)..(((((((((...))))))))).)))))..))))))).- ( -28.90, z-score = -1.55, R) >droWil1.scaffold_180955 148568 79 - 2875958 UCCCUAGAGAGUUGAAUUAGUUGAAUAAUA-------------------------UACUUAUUAUAUACAACUGAUUCUUAGUUGGGAAGUUGACAUUAUCUUU ((((.........((((((((((.((((((-------------------------....))))))...))))))))))......))))................ ( -18.06, z-score = -2.16, R) >droVir3.scaffold_13049 11678885 88 + 25233164 CGCCUAGAGAGUUGAGUCAGUGGAG----GAGGGAGGUAAAAGUCCUGAAAUAUGU----------ACCG-CUGAUUCCUAGUUGGAGAAUUGACAUAUAUAU- ...((((..((..((((((((((..----..((((........)))).........----------.)))-)))))))))..)))).................- ( -19.04, z-score = -0.47, R) >droMoj3.scaffold_6680 13786485 91 - 24764193 CGCCUAGAGAGUUGAGCCAGCGGAGC----AGGUGGGCGUUAAUUUCCCCUUGUACAA--------ACCG-CUGGGUCCUAGUUGGAGAAUUGACAAAUUAUCU ...((((..((..((.(((((((.((----(((.(((.(.....).))))))))....--------.)))-)))).))))..)))).................. ( -27.00, z-score = -0.71, R) >droGri2.scaffold_15110 5587849 102 + 24565398 CGCCUAGAGAGUUGAGCCAGUGGAACGUGGAAAGAGGAGUAUAUUAUUCCUUGGACCACCACAUCCCCCG-CUGUCUCUUAGUUGGAGAGUUGACAUUUUUUU- ..........((..(..((((((...((((...((((((((...))))))))...))))........)))-)))(((((.....))))).)..))........- ( -27.00, z-score = -0.96, R) >consensus UCCCUAGAGAGUUGAAUUACCUGAGUGGGGGGAG__G___GACUUGU___U__U_UCCCCCUGCAAACCAUCUAAUUCCUAGUUGGGAAGGUUACAUUUUCUU_ ...((((..((..((((((.......((((............................))))..........))))))))..)))).................. ( -4.64 = -3.77 + -0.87)
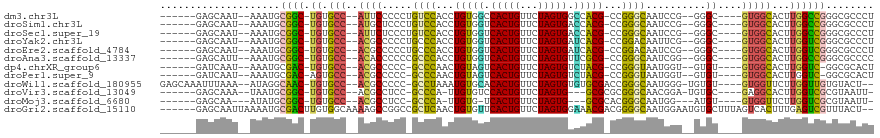
| Location | 18,889,387 – 18,889,488 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.49395 |
| G+C content | 0.59727 |
| Mean single sequence MFE | -46.67 |
| Consensus MFE | -19.95 |
| Energy contribution | -18.40 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.81 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.994041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18889387 101 - 24543557 ------GAGCAAU--AAAUGCGGC-UGUGCC--AUUCCCCCUGUCCACCUGUGGCCACUGUUCUAGUGGCCACG-CCGGGCAAUCCG--GGGC----GUGGCACUUGGCCGGGCGCCCU ------..(((..--...)))(((-((((((--((.((((.(((((...(((((((((((...)))))))))))-..)))))....)--))).----)))))))..))))((....)). ( -54.30, z-score = -3.19, R) >droSim1.chr3L 18194930 101 - 22553184 ------GAGCAAU--AAAUGCGGC-UGUGCC--AUGCUCCCUGUCCACCUGUGGUCACUGUUCUAGUGACCACG-CCGGGCAAUCCG--GGGC----GUGGCACUUGGCCGGGCGCCCU ------..(((..--...)))(((-((((((--(((((((.(((((...(((((((((((...)))))))))))-..)))))....)--))))----)))))))..))))((....)). ( -55.10, z-score = -4.20, R) >droSec1.super_19 855008 101 - 1257711 ------GAGCAAU--AAAUGCGGC-UGUGCC--AUUCUCCCUGUCCACCUGUGGUCACUGUUCUAGUGACCACG-CCGGGCAAUCCG--GGGC----GUGGCACUUGGCCGGGCGCCCU ------..(((..--...)))(((-((((((--((.((((.(((((...(((((((((((...)))))))))))-..)))))....)--))).----)))))))..))))((....)). ( -49.20, z-score = -3.03, R) >droYak2.chr3L 2448005 101 + 24197627 ------GAGCAAU--AAAUGCGGC-UGUGCC--ACGCCCCCUGCCCACCUGUGGUCACUGUUCUAGUGAUCACG-CCGGACAAUUCG--GGGC----GUGGCACUUGGUCGGGCGCCCU ------..(((..--...)))(((-((((((--(((((((.((.((...(((((((((((...)))))))))))-..)).))....)--))))----)))))))..))))((....)). ( -50.20, z-score = -3.41, R) >droEre2.scaffold_4784 18705787 101 - 25762168 ------GAGCAAU--AAAUGCGGC-UGUGCC--ACGCCCCCUGCCCACCUGUGGUCACUGUUCUAGUGAUCACG-CCGGACAAUCCG--GGGC----GUGGCACUUGGUCGGGCGCCCU ------..(((..--...)))(((-((((((--(((((((..........((((((((((...)))))))))).-..((.....)))--))))----)))))))..))))((....)). ( -50.80, z-score = -3.43, R) >droAna3.scaffold_13337 4742238 101 + 23293914 ------GAGCAUU--AAAUGCGGC-UGUGCC--ACACCCCCCGCCCACCUGUGGUCACUGUUCUAGUGUUCGCG-CCGGGCAAUCGG--GGGC----GUGGCACUUGGCCGGGCGCCCC ------..((((.--..))))(((-((((((--((.(((((.((((...((((..(((((...)))))..))))-..))))....))--))).----)))))))..))))((....)). ( -51.10, z-score = -1.98, R) >dp4.chrXR_group6 9246522 99 + 13314419 ------GAUCAAU--AAAUGCGAC-UGUGCC--ACGCCCCC-GCCCAACUGUAGUCACUGUUCUAGUGUCUACG-CCGGGUAAUGGU--GUGU----GUGGCACUUGGUC-GGCGCACU ------.......--.....((((-((((((--(((((((.-((((...(((((.(((((...))))).)))))-..))))...)).--).))----)))))))..))))-)....... ( -40.40, z-score = -2.71, R) >droPer1.super_9 1280063 99 + 3637205 ------GAUCAAU--AAAUGCGAC-AGUGCC--ACGCCCCC-GCCCAACUGUAGUCACUGUUCUAGUGUCUACG-CCGGGUAAUGGU--GUGU----GUGGCACUUGGUC-GGCGCACU ------.......--.....((((-((((((--(((((((.-((((...(((((.(((((...))))).)))))-..))))...)).--).))----))))))))..)))-)....... ( -40.40, z-score = -2.57, R) >droWil1.scaffold_180955 148647 106 - 2875958 GAGCAAAUUUAAA--AUAGGCAAC-UGUGCC--ACGCCCCC-GCCUAAAUGUGCACACUGUUCUAGUGUGUGCGACCGGGCAAUGGG-UGUGU----GUGGUUCUUGGUUGUGUACU-- .............--.((.(((((-((.(((--((((((((-((((...((..(((((((...)))))))..))...))))...)))-.).))----)))))...))))))).))..-- ( -42.40, z-score = -2.98, R) >droVir3.scaffold_13049 11678973 97 + 25233164 ------GAGCAAA--UAAUGCGGC-UGUGCC--ACGCCUCC-GCCCA-UUGUGUCCACUGUUCUAGUG---GCGCGCGGGCAACGGA-UGUGC----GAGGCACUUGGUCGCGUAAUU- ------.......--..(((((((-((((((--.(((((((-((((.-.((((.((((((...)))))---))))).))))...)))-.).))----).)))))..))))))))....- ( -45.00, z-score = -2.69, R) >droMoj3.scaffold_6680 13786576 93 - 24764193 ------GAGCAA---AUAUGCGGC-UGUGCC--ACGCCUCC-GCCCA-UUGUG-UCACUGUUCUAGUG---GCGCACGGGCAAUGG---AUGU----GUGGUUCUUGGUCGCGUAAUU- ------......---.((((((((-((.(((--((((.(((-((((.-.((((-((((((...)))))---))))).))))...))---).))----)))))...))))))))))...- ( -47.20, z-score = -4.92, R) >droGri2.scaffold_15110 5587951 111 + 24565398 ------GAGCAAUUAAAAUGCGACUUGUGGCAAAAGCCGGCCGCUCAACUGUGUUCACUGUUCUAGUGGAAACGACGGGGCAAUGGAAUGUGCUUUAGUCACUUUGAGUCGUUUACU-- ------.............((((((((((((.(((((((..(((......)))((((.((((((..((....))..)))))).)))).)).))))).)))))...))))))).....-- ( -33.90, z-score = -1.34, R) >consensus ______GAGCAAU__AAAUGCGGC_UGUGCC__ACGCCCCC_GCCCACCUGUGGUCACUGUUCUAGUGACCACG_CCGGGCAAUGGG__GGGC____GUGGCACUUGGUCGGGCGCCCU ..........................((((............((((...(((((.(((((...))))).)))))...))))..................(((.....)))..))))... (-19.95 = -18.40 + -1.55)

| Location | 18,889,458 – 18,889,553 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Shannon entropy | 0.44521 |
| G+C content | 0.47696 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -15.28 |
| Energy contribution | -14.38 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
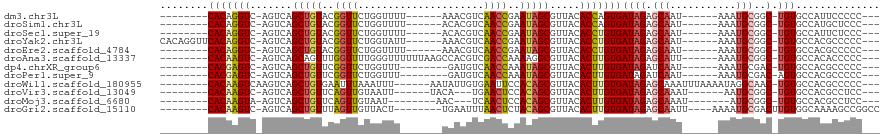
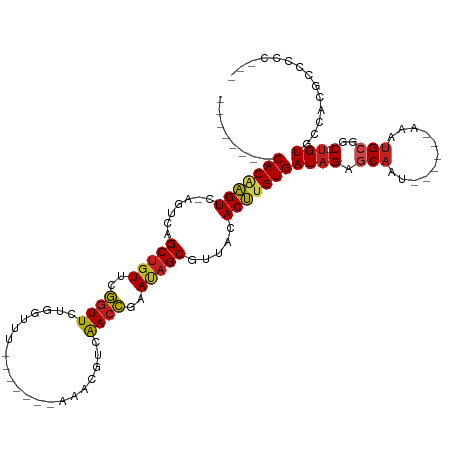
>dm3.chr3L 18889458 95 - 24543557 --------CACAGGUC-AGUCAGCUGUACGGUUCUGGUUUU------AAACGUCAACCGAAUAGCGUUACACCAGUGAUAGAGCAAU------AAAUGCGGC-UGUGCCAUUCCCCC--- --------....((((-((((.(((((.(((((.((.....------...))..))))).)))))(((((....)))))...(((..------...))))))-)).)))........--- ( -27.30, z-score = -1.76, R) >droSim1.chr3L 18195001 95 - 22553184 --------CACAGGUC-AGUCAGCUGUACGGUUCUGGUUUU------ACACGUCAACCGAAUAGCGUUACACCAGUGAUAGAGCAAU------AAAUGCGGC-UGUGCCAUGCUCCC--- --------....((((-((((.(((((.(((((.((.....------...))..))))).)))))(((((....)))))...(((..------...))))))-)).)))........--- ( -27.30, z-score = -0.90, R) >droSec1.super_19 855079 95 - 1257711 --------CACAGGUC-AGUCAGCUGUACGGUUCUGGUUUU------ACACGUCAACCGAAUAGCGUUACACCUGUGAUAGAGCAAU------AAAUGCGGC-UGUGCCAUUCUCCC--- --------(((((((.-.....(((((.(((((.((.....------...))..))))).))))).....)))))))..((((((..------...)))(((-...)))..)))...--- ( -28.10, z-score = -1.56, R) >droYak2.chr3L 2448076 103 + 24197627 CACAGGUUCACAGGUC-AGUCAGCUGUACGGUUCUGGUAUU------AAACGUCAACCGAAUAGCGUUACACCUGUGAUAGAGCAAU------AAAUGCGGC-UGUGCCACGCCCCC--- (((((..((((((((.-.....(((((.(((((.((.....------...))..))))).))))).....))))))))....(((..------...)))..)-))))..........--- ( -32.30, z-score = -1.66, R) >droEre2.scaffold_4784 18705858 95 - 25762168 --------CACAGGUC-AGUCAGCUGUACGGUUCUGGUUUU------AAACGUCAACCGAAUAGCGUUACACCUGUGAUAGAGCAAU------AAAUGCGGC-UGUGCCACGCCCCC--- --------(((((((.-.....(((((.(((((.((.....------...))..))))).))))).....))))))).....(((..------...)))(((-........)))...--- ( -28.50, z-score = -1.33, R) >droAna3.scaffold_13337 4742309 101 + 23293914 --------CACAAGUC-AGUCAGCAGUUUGGUUUUGGGUUUUUUAAGCCACGUCGACCAAAAGGCGUUACACUUGUGAUAGAGCAUU------AAAUGCGGC-UGUGCCACACCCCC--- --------(((((((.-.....((..(((((((.((((((.....)))).))..)))))))..)).....))))))).....((((.------..))))(((-...)))........--- ( -28.40, z-score = -1.13, R) >dp4.chrXR_group6 9246591 93 + 13314419 --------CACGAGUC-AGUCAGCUGUUCGGUUCUGGUUU--------GAUGUCAACCAAAUAGCGUUACACUUGUGAUAGAUCAAU------AAAUGCGAC-UGUGCCACGCCCCC--- --------...(.(((-((((.((((((.(((..(((...--------....)))))).))))))(((((....)))))........------......)))-)).)))........--- ( -19.90, z-score = 0.41, R) >droPer1.super_9 1280132 93 + 3637205 --------CACGAGUC-AGUCAGCUGUUCGGUUCUGGUUU--------GAUGUCAACCAAAUAGCGUUACACUUGUGAUAGAUCAAU------AAAUGCGAC-AGUGCCACGCCCCC--- --------(((((((.-.....((((((.(((..(((...--------....)))))).)))))).....)))))))..........------....(((..-.......)))....--- ( -18.50, z-score = 0.78, R) >droWil1.scaffold_180955 148717 102 - 2875958 --------CACAAGUCAAGUCAGCUGUGAAUUUAAAUUU------AAUAUUGUGAAUUCCACAGCGUUACACUUGUGAUAGAGCAAAUUUAAAAUAGGCAAC-UGUGCCACGCCCCC--- --------(((((((.......(((((((((((((((..------...))).)))))).)))))).....)))))))...................((((..-..))))........--- ( -24.80, z-score = -2.55, R) >droVir3.scaffold_13049 11679040 92 + 25233164 --------CACAAGUC-AGUCAGCUGUUCAGUUGUAAUU------UACA---UGAACUCCACAGCGUUACACUUGUGAUAGAGCAAAU------AAUGCGGC-UGUGCCACGCCUCC--- --------(((((((.-.....(((((..(((((((...------))))---...)))..))))).....))))))).....(((...------..)))(((-........)))...--- ( -22.60, z-score = -0.59, R) >droMoj3.scaffold_6680 13786640 89 - 24764193 --------CACAAGUA-AGUCAGCUGUUCAGUUGUAAU--------AAC---UCAACUCCACAGCGUUACACUUGUGAUAGAGCAAAU-------AUGCGGC-UGUGCCACGCCUCC--- --------(((((((.-.....(((((..(((((....--------...---.)))))..))))).....))))))).....(((...-------.)))(((-........)))...--- ( -26.30, z-score = -2.30, R) >droGri2.scaffold_15110 5588026 99 + 24565398 --------CACAAGUC-AGUCAGCUGUUUAGUUGUUACU--------UGAAUUUAACUCUACAGCGUUACACUUGUGAUAGAGCAAUU----AAAAUGCGACUUGUGGCAAAAGCCGGCC --------.(((((((-..((.(((((..(((((.....--------......)))))..)))))(((((....))))).))(((...----....))))))))))(((....))).... ( -29.50, z-score = -2.35, R) >consensus ________CACAAGUC_AGUCAGCUGUUCGGUUCUGGUUU_______AAACGUCAACCGAAUAGCGUUACACUUGUGAUAGAGCAAU______AAAUGCGGC_UGUGCCACGCCCCC___ ........(((((((.......(((((..((((.....................))))..))))).....)))))))........................................... (-15.28 = -14.38 + -0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:37 2011