| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,656,243 – 5,656,345 |
| Length | 102 |
| Max. P | 0.943333 |

| Location | 5,656,243 – 5,656,345 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.41 |
| Shannon entropy | 0.56458 |
| G+C content | 0.49208 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -11.43 |
| Energy contribution | -10.97 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.943333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
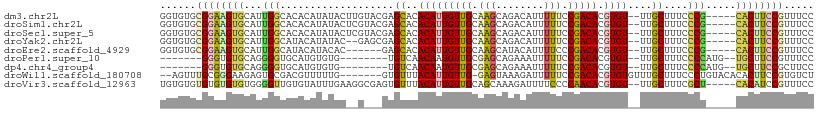
>dm3.chr2L 5656243 102 + 23011544 GGUGUGCGGAAGUGCAUUGGCACACAUAUACUUGUACGAGCACACAUUGUUGCAAGCAGACAUUUUUCCGACACGUGU--UUGCUUUCCCG-----CACUUCCGUUUCC ((...((((((((((....(((.(((......(((....))).....))))))((((((((((...........))))--))))))....)-----)))))))))..)) ( -34.20, z-score = -3.00, R) >droSim1.chr2L 5449314 102 + 22036055 GGUGUGCGGAAGUGCAUUGGCACACAUAUACUCGUACGAGCACACAUUGUUGCAAGCAGACAUUUUUCCGACACGUGU--UUGCUUUCCCG-----CACUUCCGUUUCC ((...((((((((((....(((.(((....(((....))).......))))))((((((((((...........))))--))))))....)-----)))))))))..)) ( -35.30, z-score = -3.39, R) >droSec1.super_5 3734986 102 + 5866729 GGUGUGCGGAAGUGCAUUGGCACACAUAUACUCGUACGAGCACACAUUGUUGCAAGCAGACAUUUUUCCGACACGUGU--UUGCUUUCCCG-----CACUUCCGUUUCC ((...((((((((((....(((.(((....(((....))).......))))))((((((((((...........))))--))))))....)-----)))))))))..)) ( -35.30, z-score = -3.39, R) >droYak2.chr2L 8787732 100 - 22324452 GGUGUGCGGAAGUGCAUUGGCAUACAUAUAC--GAGCGAGCACACAUUGUUGCAAGCAGACAUUUUUCCGACACGUGU--UUGCUUUCCCG-----CACUUCCGUUUCC ((...((((((((((..((.....)).....--((((((((((....(((((.(((........))).))))).))))--))))))....)-----)))))))))..)) ( -34.40, z-score = -3.00, R) >droEre2.scaffold_4929 5744626 96 + 26641161 GGUGUGCGGAAGUGCAUUGGCAUACAUACAC------GAGCACACAUUGUUGCAAGCAUACAUUUUCCCGACACGUGU--UUGCUUUCCCG-----CACUUCCGUUUCC ((...((((((((((..((.....)).....------(((((.(((((((((..((........))..))))).))))--.)))))....)-----)))))))))..)) ( -30.00, z-score = -2.36, R) >droPer1.super_10 1249527 90 - 3432795 -------GGGUGUGCAGGGGUGCAUGUGUG--------UGUCAACAAUGUUGCGAGCAGAAAUUUUUCCGACACGUGU--UUGCUUUCCCCAUG--UGCUUCCGUUUCC -------(((.(..((((((.(((.(..((--------((((.....(((.....)))(((....))).))))))..)--.)))...)))).))--..).)))...... ( -32.10, z-score = -2.37, R) >dp4.chr4_group4 3422374 90 - 6586962 -------GGGUGUGCAGGGGUGCAUGUGUG--------UGUCAACAAUGUUGCGAGCAGAAAUUUUUCCGACACGUGU--UUGCUUUCCCCAUG--UGCUUCCGCUUCC -------(((.(..((((((.(((.(..((--------((((.....(((.....)))(((....))).))))))..)--.)))...)))).))--..).)))...... ( -32.10, z-score = -2.03, R) >droWil1.scaffold_180708 3956801 99 - 12563649 --AGUUUGCGGGAAGAGUGCGACGUUUUUG-------GUGUUUACAUUGUUG-GAGUAAAGAUUUUUCCGACACGUGUGUUUGCUUUCCCUGUACACACUUCCGUGUCU --.....((((((((.(((..(((.....(-------(((..((((((((((-(((.........)))))))).)))))..)))).....))).))).))))).))).. ( -27.80, z-score = -1.34, R) >droVir3.scaffold_12963 3646572 102 + 20206255 UGUGUGUGUGUGUGUGGGGUUGUGUAUUUGAAGGCGAGUGUUUACAUUGUUGCAGCAAAGAUUUUCCCCAACACGUGU--UUGCUUUCGCU-----CACAUCCGUUUCC .(((((.(((.((((((((.......((((...(((((((....)))..))))..))))......)))).))))((..--..))...))).-----)))))........ ( -27.12, z-score = -0.69, R) >consensus GGUGUGCGGAAGUGCAUUGGCACACAUAUAC__G___GAGCACACAUUGUUGCAAGCAGACAUUUUUCCGACACGUGU__UUGCUUUCCCG_____CACUUCCGUUUCC ......((((((((...(((...................((..(((((((((.(((........))).))))).))))....))....))).....))))))))..... (-11.43 = -10.97 + -0.46)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:38 2011