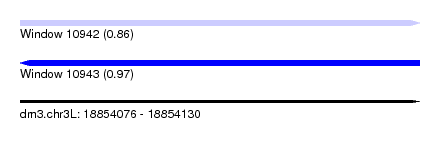
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,854,076 – 18,854,130 |
| Length | 54 |
| Max. P | 0.970160 |

| Location | 18,854,076 – 18,854,130 |
|---|---|
| Length | 54 |
| Sequences | 14 |
| Columns | 57 |
| Reading direction | forward |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.39394 |
| G+C content | 0.35719 |
| Mean single sequence MFE | -9.52 |
| Consensus MFE | -8.26 |
| Energy contribution | -8.33 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859190 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
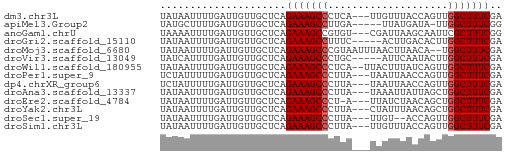
>dm3.chr3L 18854076 54 + 24543557 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUCA---UUGUUUACCAGUUGGCUUUCGA .....................(((((((...(---(((.....)))).))))))).. ( -11.00, z-score = -1.64, R) >apiMel3.Group2 8429416 51 - 14465785 UAUGCUUUUGAUUGUUGCUCAGAAAGCCUUGA-----UUAUGAUA-UUGGCUUUCGG .....................(((((((....-----........-..))))))).. ( -10.04, z-score = -0.97, R) >anoGam1.chrU 28635747 54 + 59568033 UAAAAUUUUGAUUGUUGCUCAGAAAGCCGUGU---CGAUUAAGCAAUUCGCUUUCGG .....................((((((..(((---.......)))....)))))).. ( -7.90, z-score = 0.44, R) >droGri2.scaffold_15110 429824 52 - 24565398 UAUAAUUUUGAUUGUUGCUCAGAAAGCCUUUC-----ACUUGACACUUGGCUUUCGA .....................(((((((....-----...........))))))).. ( -9.96, z-score = -1.58, R) >droMoj3.scaffold_6680 1115545 55 - 24764193 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCGUAAUUUAACUUAACA--UGGCUUUCGA .....................(((((((..(((......)))...--.))))))).. ( -9.40, z-score = -1.38, R) >droVir3.scaffold_13049 7149499 52 - 25233164 UAUCAUUUUGAUUGUUGCUCAGAAAGCCCUGC-----AUUCAAUACUUGGCUUUCGA .....................(((((((..(.-----........)..))))))).. ( -9.10, z-score = -0.61, R) >droWil1.scaffold_180955 104855 55 + 2875958 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUCA--UUACUUUAUCAGUUGGCUUUCGA .....................(((((((..(.--...........)..))))))).. ( -9.50, z-score = -1.12, R) >droPer1.super_9 1244066 54 - 3637205 UCUAUUUUUGAUUGUUGCUCAGAAAGCCCUUA---UAAUUAACCAGUUGGCUUUCGA .....................(((((((....---.............))))))).. ( -8.63, z-score = -0.90, R) >dp4.chrXR_group6 9210488 54 - 13314419 UCUAUUUUUGAUUGUUGCUCAGAAAGCCCUUA---UAAUUAACCAGUUGGCUUUCGA .....................(((((((....---.............))))))).. ( -8.63, z-score = -0.90, R) >droAna3.scaffold_13337 4708353 54 - 23293914 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUA---UAAAUUAUUAGCUGGCUUUCGA .....................(((((((....---(((....)))...))))))).. ( -9.50, z-score = -0.99, R) >droEre2.scaffold_4784 18670615 53 + 25762168 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCU-A---UUAUCUAACAGCUGGCUUUCGA .....................(((((((..-.---.............))))))).. ( -8.69, z-score = -0.57, R) >droYak2.chr3L 2410521 54 - 24197627 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUA---CUAUUUAACAGCUGGCUUUCGA .....................(((((((....---.............))))))).. ( -8.63, z-score = -0.67, R) >droSec1.super_19 820247 52 + 1257711 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUA---UUGU--ACCAGUUGGCUUUCGA .....................(((((((...(---(((.--..)))).))))))).. ( -10.60, z-score = -1.70, R) >droSim1.chr3L 18159384 54 + 22553184 UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUA---UUGUUUACCAGUUGGCUUUCGA .....................(((((((...(---(((.....)))).))))))).. ( -11.70, z-score = -2.25, R) >consensus UAUAAUUUUGAUUGUUGCUCAGAAAGCCCUUA___UAAUUUAACAGUUGGCUUUCGA .....................(((((((....................))))))).. ( -8.26 = -8.33 + 0.07)
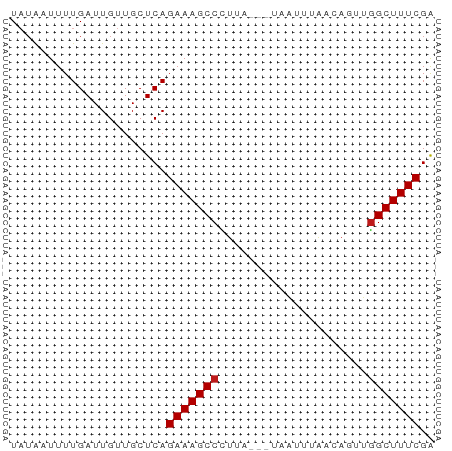
| Location | 18,854,076 – 18,854,130 |
|---|---|
| Length | 54 |
| Sequences | 14 |
| Columns | 57 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Shannon entropy | 0.39394 |
| G+C content | 0.35719 |
| Mean single sequence MFE | -10.30 |
| Consensus MFE | -8.36 |
| Energy contribution | -8.43 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.970160 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
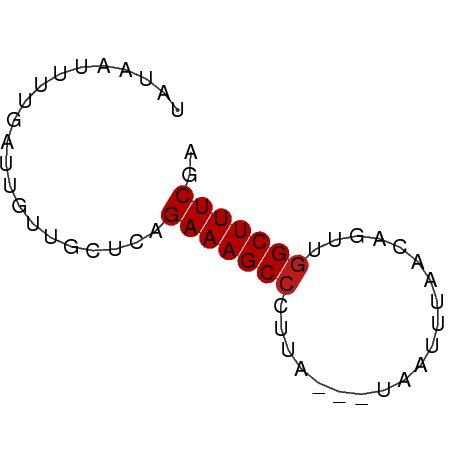
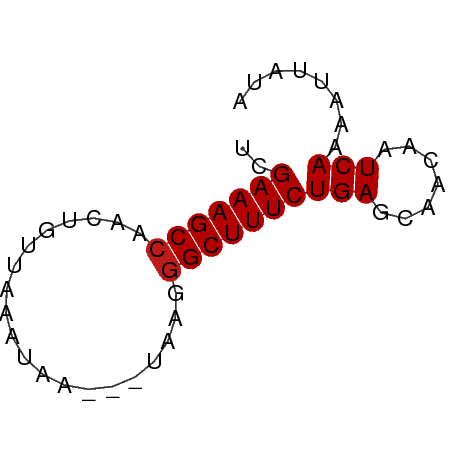
>dm3.chr3L 18854076 54 - 24543557 UCGAAAGCCAACUGGUAAACAA---UGAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((.(.((.....)).---)...)))))))(((.......)))........ ( -10.60, z-score = -1.77, R) >apiMel3.Group2 8429416 51 + 14465785 CCGAAAGCCAA-UAUCAUAA-----UCAAGGCUUUCUGAGCAACAAUCAAAAGCAUA ..(((((((..-........-----....)))))))...((...........))... ( -9.34, z-score = -2.54, R) >anoGam1.chrU 28635747 54 - 59568033 CCGAAAGCGAAUUGCUUAAUCG---ACACGGCUUUCUGAGCAACAAUCAAAAUUUUA .(((((((.....))))..)))---.....((((...))))................ ( -6.60, z-score = 0.11, R) >droGri2.scaffold_15110 429824 52 + 24565398 UCGAAAGCCAAGUGUCAAGU-----GAAAGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((.....((....-----))..)))))))(((.......)))........ ( -9.90, z-score = -1.56, R) >droMoj3.scaffold_6680 1115545 55 + 24764193 UCGAAAGCCA--UGUUAAGUUAAAUUACGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((.--(((...........))))))))))(((.......)))........ ( -10.70, z-score = -2.16, R) >droVir3.scaffold_13049 7149499 52 + 25233164 UCGAAAGCCAAGUAUUGAAU-----GCAGGGCUUUCUGAGCAACAAUCAAAAUGAUA ..(((((((..((((...))-----))..))))))).........(((.....))). ( -12.80, z-score = -2.05, R) >droWil1.scaffold_180955 104855 55 - 2875958 UCGAAAGCCAACUGAUAAAGUAA--UGAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((..((.((......)--).)))))))))(((.......)))........ ( -10.50, z-score = -1.79, R) >droPer1.super_9 1244066 54 + 3637205 UCGAAAGCCAACUGGUUAAUUA---UAAGGGCUUUCUGAGCAACAAUCAAAAAUAGA ..(((((((.....((.....)---)...)))))))(((.......)))........ ( -9.40, z-score = -1.25, R) >dp4.chrXR_group6 9210488 54 + 13314419 UCGAAAGCCAACUGGUUAAUUA---UAAGGGCUUUCUGAGCAACAAUCAAAAAUAGA ..(((((((.....((.....)---)...)))))))(((.......)))........ ( -9.40, z-score = -1.25, R) >droAna3.scaffold_13337 4708353 54 + 23293914 UCGAAAGCCAGCUAAUAAUUUA---UAAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((...(((....)))---....)))))))(((.......)))........ ( -10.00, z-score = -1.81, R) >droEre2.scaffold_4784 18670615 53 - 25762168 UCGAAAGCCAGCUGUUAGAUAA---U-AGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((..(((((....))---)-)))))))))(((.......)))........ ( -12.10, z-score = -2.04, R) >droYak2.chr3L 2410521 54 + 24197627 UCGAAAGCCAGCUGUUAAAUAG---UAAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((.(((((...))))---)...)))))))(((.......)))........ ( -13.10, z-score = -2.57, R) >droSec1.super_19 820247 52 - 1257711 UCGAAAGCCAACUGGU--ACAA---UAAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((...((..--....---))..)))))))(((.......)))........ ( -9.20, z-score = -1.57, R) >droSim1.chr3L 18159384 54 - 22553184 UCGAAAGCCAACUGGUAAACAA---UAAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((...((.....)).---....)))))))(((.......)))........ ( -10.60, z-score = -2.34, R) >consensus UCGAAAGCCAACUGUUAAAUAA___UAAGGGCUUUCUGAGCAACAAUCAAAAUUAUA ..(((((((....................)))))))(((.......)))........ ( -8.36 = -8.43 + 0.07)

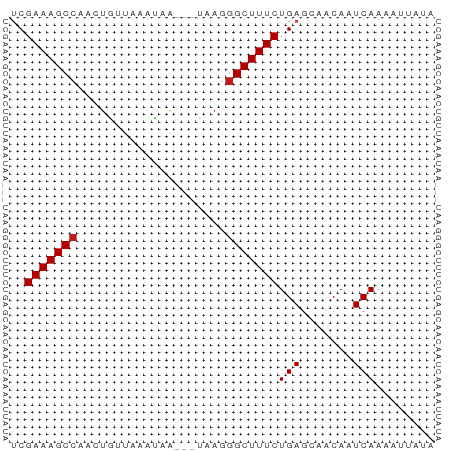
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:34 2011