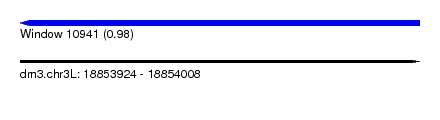
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,853,924 – 18,854,008 |
| Length | 84 |
| Max. P | 0.979068 |

| Location | 18,853,924 – 18,854,008 |
|---|---|
| Length | 84 |
| Sequences | 10 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 59.19 |
| Shannon entropy | 0.86463 |
| G+C content | 0.46081 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.59 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.77 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.979068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18853924 84 - 24543557 --------CAUCACAAUGCACUGACCAUUUGAAUG-UUAUUCUUAUUCUAGGCAGGCCAACACGUCUGCUCGGUGUGAGAGCCGCACCGGAAC --------................((....(((((-.......)))))..(((((((......))))))).((((((.....))))))))... ( -22.20, z-score = -0.87, R) >droGri2.scaffold_15110 429652 86 + 24565398 ----AUUGUAUUAUAUUUCUUUUUGCAACUGUUUGCUGAGUUUCUGUUGGC-AAAGC--AAACGUCUGCACGGUGUGAGUGCUGCACUCGAAC ----............(((....((((..((((((((..(((......)))-..)))--)))))..))))......(((((...)))))))). ( -21.10, z-score = -0.39, R) >droVir3.scaffold_13049 7149335 77 + 25233164 ----------------CAUUUUACUGUAUUGUUUGGUUGGUUUUAUUUAUGUCAAAACAACACGUCUGCACGGUGUGAGUGCUGCACUCGAAC ----------------.....((((((...(....((((.(((.((....)).))).))))....)...))))))((((((...))))))... ( -12.50, z-score = 1.39, R) >droWil1.scaffold_180955 104633 90 - 2875958 AAACAAAAGACCCAAACA-AUUGUUAGAUUGUUGGGUUA--UUAAUUUGGCUCAGAUAGCCAUGUCUGAACGGUAUGAGAGCCGGACAGGAAC ........(((((((.((-(((....)))))))))))).--......(((((.....)))))..((((..((((......))))..))))... ( -28.20, z-score = -3.80, R) >droAna3.scaffold_13337 4708193 90 + 23293914 UCACAAGAGUUAAGAAUUCUUUUUAAGACUAUUUA--GGGUUUUAUUUAGC-CAGGCCAACACGUCUGCUCGGUGUGAGAGCCGCACCGGAAC ....((((((.....))))))..(((((((.....--.)))))))...(((-.((((......)))))))(((((((.....))))))).... ( -22.30, z-score = -1.07, R) >droEre2.scaffold_4784 18670469 78 - 25762168 --------AAUCACAAUGCACUAGCUUUUGGAA-------CUCUACUCUAGGCAGGCCAACACGUCUGUUCGGUGUGAGAGCCACACCGGAAC --------............((((....(((..-------..)))..))))((((((......))))))((((((((.....))))))))... ( -21.60, z-score = -1.07, R) >droYak2.chr3L 2410376 78 + 24197627 --------AAUCACAAUGCACUAGCUGUUGGAA-------UUCUACUCUAGGCAGGCCAACACGUCUGCUCGAUGUGAGAGCCGCACCGGAAC --------..((.((((((....)).)))))).-------......((..(((.(((...((((((.....))))))...))))).)..)).. ( -19.60, z-score = -0.09, R) >droSec1.super_19 820095 84 - 1257711 --------CAACACAAUGCACUGGCCAUUUGAAUG-UUAUUCUUAUUCUAGGCAGGCCAACACGUCUGCUCGGUGUGAGAGCCGCACCGGAAC --------............((((.........((-(..(((((((.(..(((((((......)))))))..).)))))))..)))))))... ( -23.20, z-score = -0.77, R) >droSim1.chr3L 18159231 84 - 22553184 --------CAACACAAUGCACUGGCCAUUUGAAUG-UUAUUCUUAUUCUAGGCAGGCCAACACGUCUGCUCGGUGUGAGAGCCGCACCGGAAC --------............((((.........((-(..(((((((.(..(((((((......)))))))..).)))))))..)))))))... ( -23.20, z-score = -0.77, R) >triCas2.chrUn_4 1098144 84 - 1175385 --------CAUUACAAUGCAAUUGCAAUGUGUGUC-CUAUUCUAACUCUGUGUAGAAUAACACUUUCUUUCCGUUCUAACCUAACACCGAUAC --------....(((.(((....))).)))((((.-.(((((((.(.....)))))))))))).............................. ( -12.50, z-score = -1.68, R) >consensus ________CAUCACAAUGCACUGGCCAUUUGAAUG_UUAUUUUUAUUCUAGGCAGGCCAACACGUCUGCUCGGUGUGAGAGCCGCACCGGAAC ...................................................((((((......)))))).(((((((.....))))))).... (-10.12 = -10.59 + 0.47)

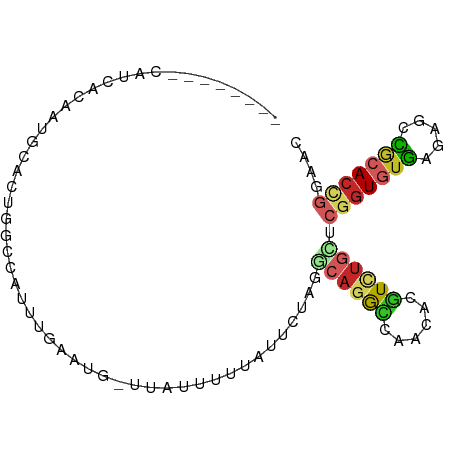
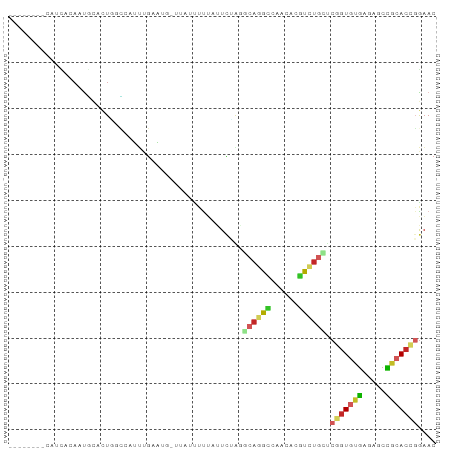
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:32 2011