| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,836,804 – 18,836,911 |
| Length | 107 |
| Max. P | 0.771254 |

| Location | 18,836,804 – 18,836,911 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 61.09 |
| Shannon entropy | 0.71000 |
| G+C content | 0.30492 |
| Mean single sequence MFE | -18.13 |
| Consensus MFE | -4.95 |
| Energy contribution | -5.06 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.771254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18836804 107 - 24543557 CACUUGAUCAACUUGCUGCGAAAAAAAAACUUUCCACAUAUGUUUCUACACACAUA-AGAUACAUUUAUGAUUCAUAU-AGCA--------UUUUCUUGCUUAAACAGCAAGAUCAA ....(((((...((((((..................(((((((.(((.........-))).)))..))))........-((((--------......))))....))))))))))). ( -18.50, z-score = -2.54, R) >dp4.chrXR_group6 9194611 83 + 13314419 CACUUGGUAUAUUUCCUAU------AAAAACAU--GGUUAU------------------AUGAUUGUGCGGCUGGU-UAUAC-------UAUACGCUUGUUUAUUCAACAAGAUCAA ....(((((((...((...------.....((.--.(((..------------------..)))..)).....)).-)))))-------))....((((((.....))))))..... ( -13.22, z-score = -0.01, R) >droAna3.scaffold_13337 4694250 86 + 23293914 CACUUGAGGCAUUUAAUAU------AAAAAAAUUCACAUAA------------------AUGGGGAUAUAGAAGAUAUAUAU------GUAUAU-CUGGCUUAUUCAGCAAGAUCAA ..((((...((((((....------.............)))------------------))).(((((....(((((((...------.)))))-))....)))))..))))..... ( -13.73, z-score = -1.14, R) >droEre2.scaffold_4784 10664169 102 + 25762168 CACUUGAUACAUAUGCUACU---AAAAAACGUUUUACAUACAUACAU------------AUAAAUGUAUGAUUCACAUGAACAUUUUCUUGUUUUCCUGUUUAAACAGCAAGAUCAG ....((((.....((((...---.................(((((((------------....)))))))....(((.(((((......)))))...)))......))))..)))). ( -15.00, z-score = -1.19, R) >droYak2.chr3L 8961049 112 + 24197627 CACUUGAUCCACUUGCUGCG-----AAAAACUGACACAUAUGUACAUCCAUAUGUACAUAUAACUGUAUGAUUCAUAUGAACAUUUUCUUGUUUUCUUGUUUAAACAGCGAGAUCUG ...........(((((((.(-----((......(((.((((((((((....))))))))))...)))....)))...((((((..............))))))..)))))))..... ( -23.24, z-score = -2.22, R) >droSec1.super_19 799321 102 - 1257711 CACUUGAUCAAUUCGCUGCG---AAAAAACUUUCCACAUAUGUAUAUACAUUCAU----AUACAUUCAUGAUUCAUGUGAAUA--------UUUUCUUGCUUAAACAGCAAGAUCAA ....(((((.((((((((.(---(((....))))))(((((((((((......))----))))))..)))......)))))).--------.....(((((.....)))))))))). ( -21.50, z-score = -3.18, R) >droSim1.chr3L 18137581 102 - 22553184 CACUUGAUCAAUUCGCUGCG---AAAAAACUUUCCACAUAUGUAUAUACAUUCAU----AUACAUGUAUGAUUCAUAUGAACA--------UUUUCUUGCUUAAAUAGCAAGAUCAA ((..((((((....(.((.(---(((....)))))))((((((((((......))----)))))))).))).)))..))....--------...(((((((.....))))))).... ( -21.70, z-score = -3.21, R) >consensus CACUUGAUCAAUUUGCUGCG___AAAAAACUUUCCACAUAUGUAUAU_CA____U____AUAAAUGUAUGAUUCAUAUGAACA______U_UUUUCUUGCUUAAACAGCAAGAUCAA ..............................................................................................(((((((.....))))))).... ( -4.95 = -5.06 + 0.10)

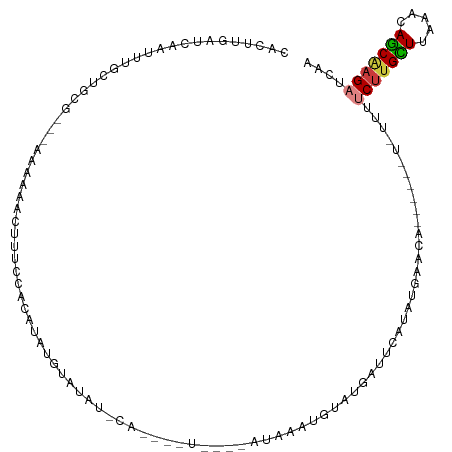

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:31 2011