| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,816,537 – 18,816,617 |
| Length | 80 |
| Max. P | 0.774442 |

| Location | 18,816,537 – 18,816,617 |
|---|---|
| Length | 80 |
| Sequences | 8 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 64.18 |
| Shannon entropy | 0.70568 |
| G+C content | 0.35983 |
| Mean single sequence MFE | -15.46 |
| Consensus MFE | -6.28 |
| Energy contribution | -5.88 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.60 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.774442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18816537 80 - 24543557 GAGGGCACGAUUCCACAAUUGAAAUCA-UGCAUUUAAUCUAAAC-------CAAA---AGCGAAAGCGAAUAAUUAACACAAAAGUGCAAA ....((((((((.((....)).)))).-................-------....---.((....)).................))))... ( -13.60, z-score = -1.99, R) >droPer1.super_9 1204406 77 + 3637205 ----------AAACUAAAUCUAAAUUA-AAAAUAUGGUGCACGUUGCUCAACAAA---AGCGAGAGCGAGUAAUUAACACAAAAGUGGUAA ----------.................-........(((...(((((((......---.((....)))))))))...)))........... ( -11.01, z-score = -0.42, R) >dp4.chrXR_group6 9171807 77 + 13314419 ----------AAACUAAAUCUAAAUUA-AAAAUAUGGUGCACGUUGCUCAACAAA---AGCGAGAGCAAGUAAUUAACACAAAAGUGGUAA ----------.................-.........(((((.((((((......---.....))))))))......(((....)))))). ( -9.90, z-score = -0.15, R) >droEre2.scaffold_4784 10646244 87 + 25762168 GAGGGCACGAUUCUUCAAUUGAAAUCA-UGCAUUUAAUCUGAGCGUUCAGGCAAA---AGCGAAAGCGAAUAAUUAACACGAAAGUGCAAA ....(((.((((..((....)))))).-)))......((((......))))....---.((....))..........(((....))).... ( -17.70, z-score = -0.59, R) >droYak2.chr3L 8942835 87 + 24197627 GAGGGCACGAUUCCUCAAUUGAAAUCA-UGCAUUUAAUCUGGGCGUUCAGGCAAA---AGCGAAAGCGAAUAAUUAACACGAAAGUGCAAA (((((......)))))...........-(((((((..(((((....)))))....---.((....))...............))))))).. ( -19.60, z-score = -1.14, R) >droSec1.super_19 780691 87 - 1257711 GAGGGCACGAUUCCUCAAUUGAAAUCG-UGUAUUUAAUCUGAGCGUUCAGCCAAA---AGCGAAAGCGAAUAAUUAACACGAAAGUGCAAA ..((((((((((.(......).)))))-))).......(((......)))))...---.((....))..........(((....))).... ( -22.90, z-score = -2.43, R) >droSim1.chr3L 18118420 87 - 22553184 GAGGGCACGAUUCCUCAAUUGAAAUCA-UGCAUUUCAUCUGAGCGUUCAGCCAAA---AGCGAAAGCGAAUAAUUAACACGAAAGUGCAAA ...(((..((...((((..((((((..-...))))))..))))...)).)))...---.((....))..........(((....))).... ( -23.30, z-score = -2.50, R) >apiMel3.GroupUn 38824211 81 - 399230636 -------UAUCAUCUAAA---GAAUUACAAAAUGUAGUCAUUCUUAACAAAGAAAGAGAAUCAAUAUCAGAAAACAAUACCGAGACAAUGA -------..((.(((...---((.((((.....)))))).(((((....))))))))))................................ ( -5.70, z-score = 0.40, R) >consensus GAGGGCACGAUUCCUCAAUUGAAAUCA_UGCAUUUAAUCUGAGCGUUCAGACAAA___AGCGAAAGCGAAUAAUUAACACGAAAGUGCAAA ...........................................................((....))..........(((....))).... ( -6.28 = -5.88 + -0.41)

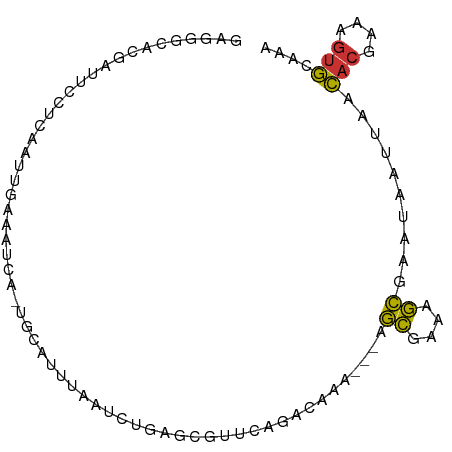
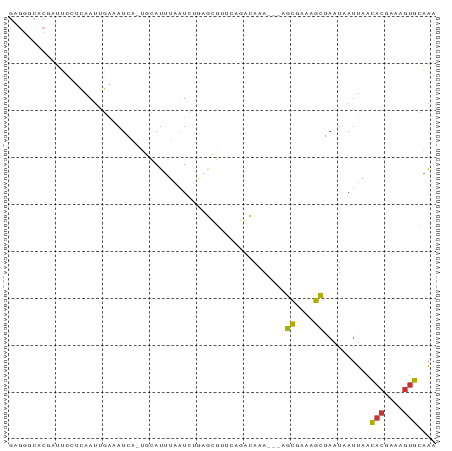
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:29 2011