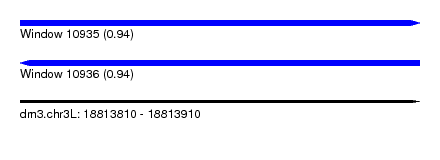
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,813,810 – 18,813,910 |
| Length | 100 |
| Max. P | 0.944832 |

| Location | 18,813,810 – 18,813,910 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.58011 |
| G+C content | 0.43452 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -14.72 |
| Energy contribution | -16.42 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
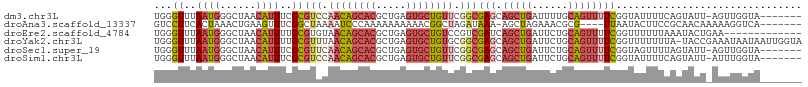
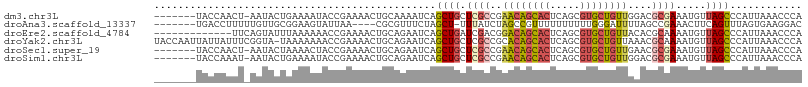
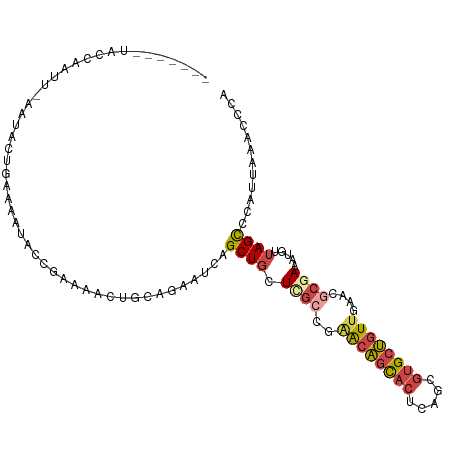
>dm3.chr3L 18813810 100 + 24543557 UGGGUUUAAUGGGCUAACAUUUCGCGUCCAACAGCACGCUGAGUGCUGUUCGGCGAGCAGCUGAUUUUGCAGUUUUCGGUAUUUUCAGUAUU-AGUUGGUA------- ............((((((......((((.((((((((.....)))))))).))))....(((((...(((........)))...)))))...-.)))))).------- ( -31.70, z-score = -2.01, R) >droAna3.scaffold_13337 4673009 96 - 23293914 GUCCUUCACUAAACUGAAGUUUCGGCUAAAAUCCCAAAAAAAAAACGGCUAGAUAAA-AGCUAGAAACGCG----UUAAUACUUCCGCAACAAAAAGGUCA------- (.((((............(((((((........))...........((((.......-)))).)))))(((----..(....)..)))......)))).).------- ( -15.40, z-score = -1.00, R) >droEre2.scaffold_4784 10643546 95 - 25762168 UGGGUUUAAUGGGCUAACAUUUUGCGUGUAACAGCACGCUGAGUGCUGUCCGUCGAUCAGCUGAUUCUGCAGUUUUCGGUUUUUUAAAUACUGAA------------- ..((((..((((((...(((((.((((((....)))))).)))))..)))))).))))(((((......)))))((((((.........))))))------------- ( -28.40, z-score = -1.83, R) >droYak2.chr3L 8939986 107 - 24197627 UGGGUUUAAUGGGCUAACAUUUUGCGUUUAACAGCACGCUGAGUGCUGUGCGGCGAGCAGCUGAUUCUGCAGUUUUCGGUUUUUUUA-UACCGAAAUAAUAAUUGGUA ............(((((....((((.....(((((((.....))))))).((((.....)))).....)))).(((((((.......-.)))))))......))))). ( -29.90, z-score = -0.93, R) >droSec1.super_19 777923 100 + 1257711 UGGGUUUAAUGGGCUAACAUUUCGCGUUCAACAGCACGCUGAGUGCUGUUCGGCGAGCAGCUGAUUCUGCAGUUUUCGGUAGUUUUAGUAUU-AGUUGGUA------- ............((((((..(((((....((((((((.....))))))))..)))))..(((((..((((........))))..)))))...-.)))))).------- ( -32.30, z-score = -2.10, R) >droSim1.chr3L 18115632 100 + 22553184 UGGGUUUAAUGGGCUAACAUUUCGCGUCCAACAGCACGCUGAGUGCUGUUCGGCGAGCAGCUGAUUCUGCAGUUUUCGGUAUUUUCAGUAUU-AUUUGGUA------- ............(((((.......((((.((((((((.....)))))))).))))....(((((...(((........)))...)))))...-..))))).------- ( -27.10, z-score = -0.48, R) >consensus UGGGUUUAAUGGGCUAACAUUUCGCGUCCAACAGCACGCUGAGUGCUGUUCGGCGAGCAGCUGAUUCUGCAGUUUUCGGUAUUUUCAGUACU_AAUUGGUA_______ ...((..((((......))))..))(((.((((((((.....)))))))).)))(((.(((((......))))))))............................... (-14.72 = -16.42 + 1.70)
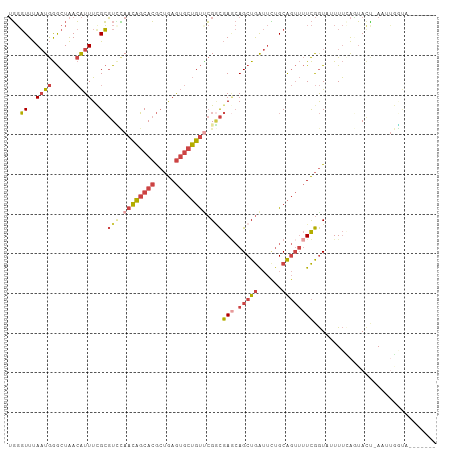
| Location | 18,813,810 – 18,813,910 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.58011 |
| G+C content | 0.43452 |
| Mean single sequence MFE | -21.77 |
| Consensus MFE | -9.97 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
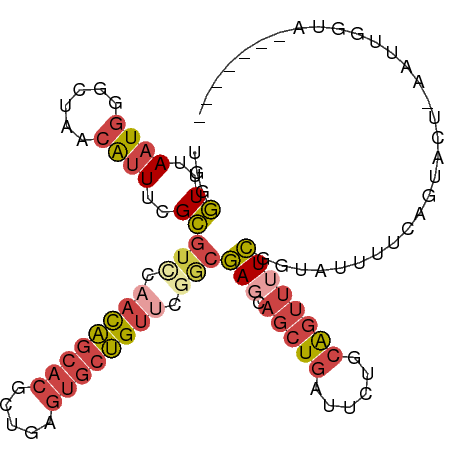
>dm3.chr3L 18813810 100 - 24543557 -------UACCAACU-AAUACUGAAAAUACCGAAAACUGCAAAAUCAGCUGCUCGCCGAACAGCACUCAGCGUGCUGUUGGACGCGAAAUGUUAGCCCAUUAAACCCA -------......((-((((..................(((........)))(((((.((((((((.....)))))))).)..))))..))))))............. ( -22.50, z-score = -2.24, R) >droAna3.scaffold_13337 4673009 96 + 23293914 -------UGACCUUUUUGUUGCGGAAGUAUUAA----CGCGUUUCUAGCU-UUUAUCUAGCCGUUUUUUUUUUGGGAUUUUAGCCGAAACUUCAGUUUAGUGAAGGAC -------...(((((.....((.(((((.....----.(((...((((..-.....)))).)))......(((((........)))))))))).)).....))))).. ( -17.70, z-score = 0.46, R) >droEre2.scaffold_4784 10643546 95 + 25762168 -------------UUCAGUAUUUAAAAAACCGAAAACUGCAGAAUCAGCUGAUCGACGGACAGCACUCAGCGUGCUGUUACACGCAAAAUGUUAGCCCAUUAAACCCA -------------..((((.(((........))).))))........((((((((...((((((((.....))))))))...))......))))))............ ( -19.40, z-score = -1.23, R) >droYak2.chr3L 8939986 107 + 24197627 UACCAAUUAUUAUUUCGGUA-UAAAAAAACCGAAAACUGCAGAAUCAGCUGCUCGCCGCACAGCACUCAGCGUGCUGUUAAACGCAAAAUGUUAGCCCAUUAAACCCA ............(((((((.-.......)))))))...((((......))))..((...(((((((.....))))))).....))..((((......))))....... ( -23.80, z-score = -2.22, R) >droSec1.super_19 777923 100 - 1257711 -------UACCAACU-AAUACUAAAACUACCGAAAACUGCAGAAUCAGCUGCUCGCCGAACAGCACUCAGCGUGCUGUUGAACGCGAAAUGUUAGCCCAUUAAACCCA -------......((-((((..................((((......))))((((..((((((((.....))))))))....))))..))))))............. ( -23.70, z-score = -3.24, R) >droSim1.chr3L 18115632 100 - 22553184 -------UACCAAAU-AAUACUGAAAAUACCGAAAACUGCAGAAUCAGCUGCUCGCCGAACAGCACUCAGCGUGCUGUUGGACGCGAAAUGUUAGCCCAUUAAACCCA -------........-....((((..............((((......))))(((((.((((((((.....)))))))).)..))))....))))............. ( -23.50, z-score = -2.32, R) >consensus _______UACCAAUU_AAUACUGAAAAUACCGAAAACUGCAGAAUCAGCUGCUCGCCGAACAGCACUCAGCGUGCUGUUGAACGCGAAAUGUUAGCCCAUUAAACCCA ...............................................((((.((((..((((((((.....))))))))....)))).....))))............ ( -9.97 = -10.92 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:28 2011