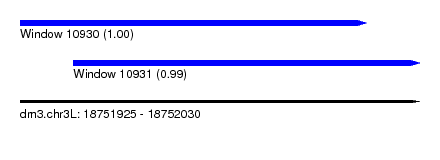
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,751,925 – 18,752,030 |
| Length | 105 |
| Max. P | 0.995590 |

| Location | 18,751,925 – 18,752,016 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 74.50 |
| Shannon entropy | 0.48395 |
| G+C content | 0.46507 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.37 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18751925 91 + 24543557 UGACCCUACCUCCGUAACAAAAAACAUAUGGCAU-UCGAGCCAUGUGAGUCAUGUU-------ACCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUAUAG .............((((((..(..((((((((..-....))))))))..)..))))-------))(((((..((.(....).--)).)))))......... ( -26.90, z-score = -4.12, R) >droSim1.chr3L 18059915 90 + 22553184 UGACCCUAACUCCGUAACAAAAA-CAUAUGGCAU-UCGAGCCAUGUGAGUCAUGUU-------ACCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUAUAG .............((((((....-((((((((..-....)))))))).....))))-------))(((((..((.(....).--)).)))))......... ( -26.50, z-score = -3.88, R) >droSec1.super_19 724989 90 + 1257711 UGACCCUAACUCCUUAACAAAAA-CAUAUGGCAU-UCGAGCCAUGUGAGUCAUGUU-------ACCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUAUAG ..............(((((....-((((((((..-....)))))))).....))))-------).(((((..((.(....).--)).)))))......... ( -23.40, z-score = -3.00, R) >droYak2.chr3L 8885086 90 - 24197627 UGACCCUACCGCCGUAACAAAAA-CAUAUGGCAU-UCGAGCCAUGUGAGUCAUGAU-------GGCCAGCUAAGCCGCAAGU--CUCGCUGGUCUUUACAG ((((..(((....))).......-((((((((..-....)))))))).))))(((.-------(((((((..((.(....).--)).))))))).)))... ( -28.90, z-score = -2.69, R) >droEre2.scaffold_4784 10589426 90 - 25762168 UGACCCUACAUCCGUAACAAAAA-CAUAUGGCAU-UCGAGCCAUGUGAGUCAUGUG-------GCCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUACAG ((((..(((....))).......-((((((((..-....)))))))).))))((((-------(.(((((..((.(....).--)).)))))...))))). ( -27.00, z-score = -2.75, R) >droAna3.scaffold_13337 4617449 98 - 23293914 UGUUUGGACCUUAGCAACAACAA-CAUAUGGCAU-AUAAGCCAUGUGAGUCAUAUCCAAUUUAACCCAGCAAAGCCGAAAGUAACGCGCUGG-CUUUAUAG ...(((((.....(......)..-((((((((..-....)))))))).......)))))......(((((.....(....)......)))))-........ ( -22.50, z-score = -1.33, R) >droVir3.scaffold_13049 7057176 82 - 25233164 UCACUCUCAUUCGAUUACAGCGG-CAGUUAUCAUGACGAGUUUGGCAAAACGUAGGC------ACUGAGCCAAACCGG---------GCUGAAGUUUA--- ............((((.((((..-((.......)).((.(((((((...........------.....))))))))).---------)))).))))..--- ( -16.39, z-score = 0.74, R) >consensus UGACCCUACCUCCGUAACAAAAA_CAUAUGGCAU_UCGAGCCAUGUGAGUCAUGUU_______ACCCAGCUAAGCCGAAAGU__CUCGCUGGUCUUUAUAG ........................((((((((.......))))))))..................(((((.....(....)......)))))......... (-15.10 = -15.37 + 0.27)
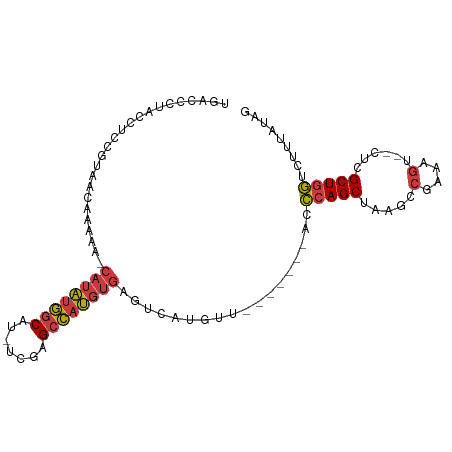
| Location | 18,751,939 – 18,752,030 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.71 |
| Shannon entropy | 0.22159 |
| G+C content | 0.44131 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -19.87 |
| Energy contribution | -19.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.987119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18751939 91 + 24543557 UAACAAAAAACAUAUGGCAUUCGAGCCAUGUGAGUCAUGUU-------ACCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUAUAGCCCCCUGAAAUAAA (((((..(..((((((((......))))))))..)..))))-------).(((((..((.(....).--)).)))))....................... ( -23.60, z-score = -3.21, R) >droSim1.chr3L 18059929 90 + 22553184 UAACAAAAA-CAUAUGGCAUUCGAGCCAUGUGAGUCAUGUU-------ACCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUAUAGCCCCCUGAAAUAAA (((((....-((((((((......)))))))).....))))-------).(((((..((.(....).--)).)))))....................... ( -23.20, z-score = -3.06, R) >droSec1.super_19 725003 90 + 1257711 UAACAAAAA-CAUAUGGCAUUCGAGCCAUGUGAGUCAUGUU-------ACCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUAUAGCCCCCUGAAAUACA (((((....-((((((((......)))))))).....))))-------).(((((..((.(....).--)).)))))....................... ( -23.20, z-score = -2.77, R) >droYak2.chr3L 8885100 90 - 24197627 UAACAAAAA-CAUAUGGCAUUCGAGCCAUGUGAGUCAUGAU-------GGCCAGCUAAGCCGCAAGU--CUCGCUGGUCUUUACAGCGCCCUGAAAUAAA .........-((((((((......)))))))).((..(((.-------(((((((..((.(....).--)).))))))).)))..))............. ( -28.50, z-score = -2.64, R) >droEre2.scaffold_4784 10589440 90 - 25762168 UAACAAAAA-CAUAUGGCAUUCGAGCCAUGUGAGUCAUGUG-------GCCCAGCUAAGCCGAAAGU--CUCGCUGGUCUUUACAGCGCCCUCAAAUAAA .........-((((((((......)))))))).((..((((-------(.(((((..((.(....).--)).)))))...)))))..))........... ( -24.40, z-score = -1.89, R) >droAna3.scaffold_13337 4617463 98 - 23293914 CAACAACAA-CAUAUGGCAUAUAAGCCAUGUGAGUCAUAUCCAAUUUAACCCAGCAAAGCCGAAAGUAACGCGCUGG-CUUUAUAGCCGCCAAAAAAAAA .........-((((((((......)))))))).....................((..(((((.......)).)))((-((....)))))).......... ( -22.70, z-score = -2.71, R) >consensus UAACAAAAA_CAUAUGGCAUUCGAGCCAUGUGAGUCAUGUU_______ACCCAGCUAAGCCGAAAGU__CUCGCUGGUCUUUAUAGCCCCCUGAAAUAAA ..........((((((((......))))))))..................(((((.....(....)......)))))....................... (-19.87 = -19.87 + -0.00)

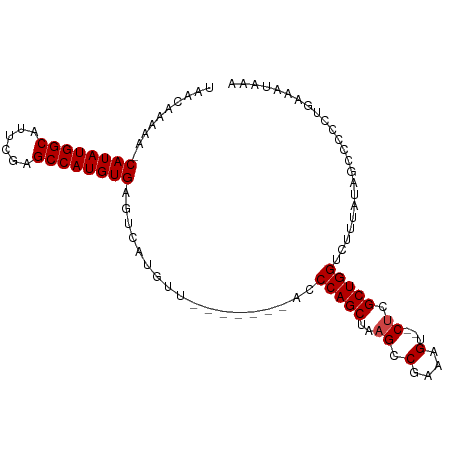
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:24 2011