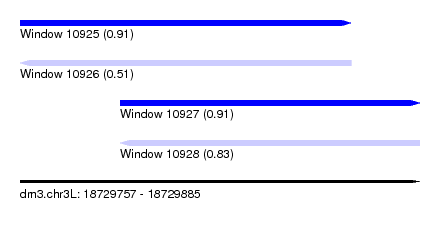
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,729,757 – 18,729,885 |
| Length | 128 |
| Max. P | 0.913304 |

| Location | 18,729,757 – 18,729,863 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.55393 |
| G+C content | 0.52838 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.70 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18729757 106 + 24543557 ---UGUACGUGCCACAAAAAGGGGGAGAUUCACCACCAGAA-GCACGAAUUUUGCAGCAGAUGCAGCCGAGAUCCC-GAAAAGGGAUCAACAGGUGGGGAAUCCAGCUGGC------- ---....(((((........((((........)).))....-))))).....((((.....))))(((..((((((-.....))))))...((.(((.....))).)))))------- ( -33.10, z-score = -1.64, R) >droSim1.chr3L 18037469 111 + 22553184 ---UGUACGUGCCACAAAAUGGGG-AGAUUC--CGCCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUGCGGAAUCCGAGUGGGAGAAUCC ---.....(..((......(((((-((((((--.((......))..)))))))(((.....)))..))))((((((-.....))))))....))..)(((.(((.....)))...))) ( -39.60, z-score = -3.04, R) >droSec1.super_19 702585 104 + 1257711 ---UGUACGUGCCACAAAAUGGGG-AGAUUC--CGCCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUGCGGAAUCCGAGUGGG------- ---....((..((......(((((-((((((--.((......))..)))))))(((.....)))..))))((((((-.....))))))....))..))....((....)).------- ( -37.30, z-score = -2.81, R) >droYak2.chr3L 8862935 111 - 24197627 ---UGUACGUGCCACAAAAUGGGG-AGAUUC--CACCAGAA-GCUGCAAAUUUGCAGCAGCUGCAGCGCGGAUCCCCAAAAGGGGAUCAACAGGUGGGGCAGCCCAGUGGGAAAAUCC ---........((((....((((.-.(.(((--((((....-((((((....)))))).((....))(..(((((((....)))))))..).))))))))..))))))))........ ( -46.70, z-score = -3.17, R) >droEre2.scaffold_4784 10567636 110 - 25762168 ---UGUACGUGCCACAAAAUGGGC-AGAUUC--CGCCAGCA-GCACGAAAUUUGCAGCAGAUGCAGCGCAGAUCCC-AAAAAGGGAUCAACAGGUUGGGCAUCCGAGUGGGAGAAUCC ---......((((........)))-)(((((--.(((..((-((.......(((((.....)))))....((((((-.....)))))).....))))))).(((.....)))))))). ( -34.00, z-score = -1.14, R) >droPer1.super_20 1527985 98 + 1872136 UGCCAUAGUUGGCACAUCCAGAGCCAGAAUC--GGCCAGAGAUCAACAGGUU-AUAGGAAAUUUAAUGGAGAUUCCCGGAAGGUCUCUACUUGAUUUAUGC----------------- (((((....)))))..(((.(.(((......--))))...((((....))))-...)))...((((((((((...((....)))))))).)))).......----------------- ( -24.80, z-score = -0.73, R) >consensus ___UGUACGUGCCACAAAAUGGGG_AGAUUC__CGCCAGAA_GCACGAAUUUUGCAGCAGAUGCAGCCCAGAUCCC_GAAAAGGGAUCAACAGGUGGGGAAUCCGAGUGGG_______ ...........((((..................((((................((.((....)).))...((((((......))))))....)))).(.....)..))))........ (-14.25 = -14.70 + 0.45)
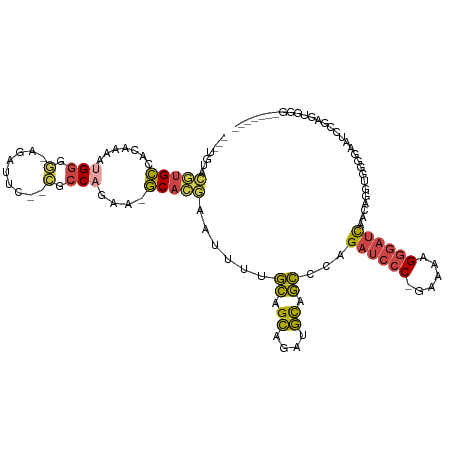
| Location | 18,729,757 – 18,729,863 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 69.42 |
| Shannon entropy | 0.55393 |
| G+C content | 0.52838 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -11.99 |
| Energy contribution | -13.41 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.507106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18729757 106 - 24543557 -------GCCAGCUGGAUUCCCCACCUGUUGAUCCCUUUUC-GGGAUCUCGGCUGCAUCUGCUGCAAAAUUCGUGC-UUCUGGUGGUGAAUCUCCCCCUUUUUGUGGCACGUACA--- -------(((((..((((((.(((((....((((((.....-))))))...((.((....)).))...........-....))))).))))))..)........)))).......--- ( -33.40, z-score = -1.99, R) >droSim1.chr3L 18037469 111 - 22553184 GGAUUCUCCCACUCGGAUUCCGCACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGGCG--GAAUCU-CCCCAUUUUGUGGCACGUACA--- (((...(((.....))).)))((((((...((((((.....-)))))).))).)))((.(((..((((((....(((....)))(--(.....-))..))))))..))).))...--- ( -36.20, z-score = -1.76, R) >droSec1.super_19 702585 104 - 1257711 -------CCCACUCGGAUUCCGCACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGGCG--GAAUCU-CCCCAUUUUGUGGCACGUACA--- -------.......(((((.(((((((...((((((.....-)))))).)))..((....)))))).)))))(((((...(((.(--(.....-)))))......))))).....--- ( -33.70, z-score = -1.69, R) >droYak2.chr3L 8862935 111 + 24197627 GGAUUUUCCCACUGGGCUGCCCCACCUGUUGAUCCCCUUUUGGGGAUCCGCGCUGCAGCUGCUGCAAAUUUGCAGC-UUCUGGUG--GAAUCU-CCCCAUUUUGUGGCACGUACA--- (((..((((.((..((((((......(((.(((((((....))))))).)))..))))).((((((....))))))-..)..)))--)))..)-))((((...))))........--- ( -45.70, z-score = -3.37, R) >droEre2.scaffold_4784 10567636 110 + 25762168 GGAUUCUCCCACUCGGAUGCCCAACCUGUUGAUCCCUUUUU-GGGAUCUGCGCUGCAUCUGCUGCAAAUUUCGUGC-UGCUGGCG--GAAUCU-GCCCAUUUUGUGGCACGUACA--- (((((((.(((...((((((......(((.((((((.....-)))))).)))..))))))((.(((.......)))-.))))).)--))))))-((((.....).))).......--- ( -37.50, z-score = -2.36, R) >droPer1.super_20 1527985 98 - 1872136 -----------------GCAUAAAUCAAGUAGAGACCUUCCGGGAAUCUCCAUUAAAUUUCCUAU-AACCUGUUGAUCUCUGGCC--GAUUCUGGCUCUGGAUGUGCCAACUAUGGCA -----------------............((((((.(...((((.....................-..))))..).))))))(((--(....((((.(.....).))))....)))). ( -19.90, z-score = 0.58, R) >consensus _______CCCACUCGGAUUCCCCACCUGUUGAUCCCUUUUC_GGGAUCUGCGCUGCAUCUGCUGCAAAAUUCGUGC_UUCUGGCG__GAAUCU_CCCCAUUUUGUGGCACGUACA___ ..............................((((((......))))))...........((((((((((........(((.......))).........))))))))))......... (-11.99 = -13.41 + 1.42)
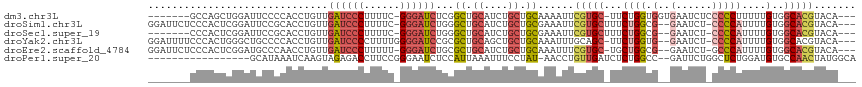

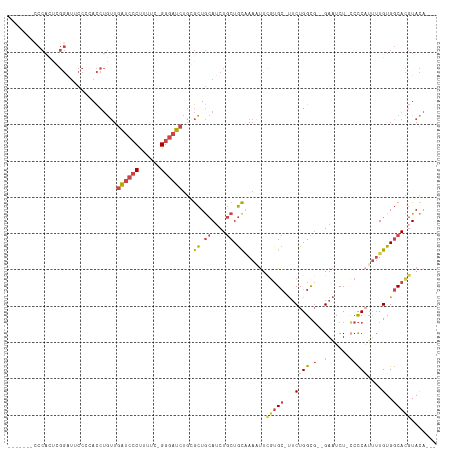
| Location | 18,729,789 – 18,729,885 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.49177 |
| G+C content | 0.55796 |
| Mean single sequence MFE | -40.34 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.26 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18729789 96 + 24543557 CCAGAA-GCACGAAUUUUGCAGCAGAUGCAGCCGAGAUCCC-GAAAAGGGAUCAACAGGUG--------------GGGAAUCCAGCUGGCGAGCAUG------UGCAUCCAUCUAGCU ..(((.-(((.......))).(((.((((.(((..((((((-.....))))))...((.((--------------(.....))).)))))..)))).------))).....))).... ( -35.50, z-score = -2.38, R) >droSim1.chr3L 18037498 111 + 22553184 CCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUGCGGAAUCCGAGUGGGAGAAUCCAGCUGGCGAGCAUG------UGCAUCCAUCUAGCU .......(((((...((((((((....(((.((..((((((-.....))))))....)))))(((.(((.....)))...))).))).)))))..))------)))............ ( -38.60, z-score = -1.89, R) >droSec1.super_19 702614 79 + 1257711 CCAGAAAGCACGAAUUUCGCAGCAGAUGCAGCCCAGAUCCC-GAAAAGGGAUCAACAGGUG--------------CGGAAUCCGAGUGG-GAGCU----------------------- ((.....((.((.(((((((.((....)).(((..((((((-.....))))))....))))--------------)))))).)).)).)-)....----------------------- ( -26.90, z-score = -2.41, R) >droYak2.chr3L 8862964 117 - 24197627 CCAGAA-GCUGCAAAUUUGCAGCAGCUGCAGCGCGGAUCCCCAAAAGGGGAUCAACAGGUGGGGCAGCCCAGUGGGAAAAUCCAGCUAGCGAGCUUUCCCGGCUGCAUCCAUCUAGUU .(((..-((((((....))))))..))).((((..(((((((....)))))))..)(((((((((((((....(((((.....((((....))))))))))))))).))))))).))) ( -55.80, z-score = -4.05, R) >droEre2.scaffold_4784 10567665 116 - 25762168 CCAGCA-GCACGAAAUUUGCAGCAGAUGCAGCGCAGAUCCC-AAAAAGGGAUCAACAGGUUGGGCAUCCGAGUGGGAGAAUCCAGCUGCCGGGUUUUCCCGGCUGCAGCCACCUAGUU ...((.-(((.......))).))...((((((((.((((((-.....))))))....(((((((..(((.....)))...)))))))))((((....))))))))))........... ( -44.90, z-score = -2.15, R) >consensus CCAGAA_GCACGAAUUUUGCAGCAGAUGCAGCCCAGAUCCC_GAAAAGGGAUCAACAGGUG_GG_A_CC_AGUGGGAGAAUCCAGCUGGCGAGCUUG______UGCAUCCAUCUAGCU .......((.........((.((....)).))...((((((......)))))).......................................))........................ (-14.42 = -14.26 + -0.16)

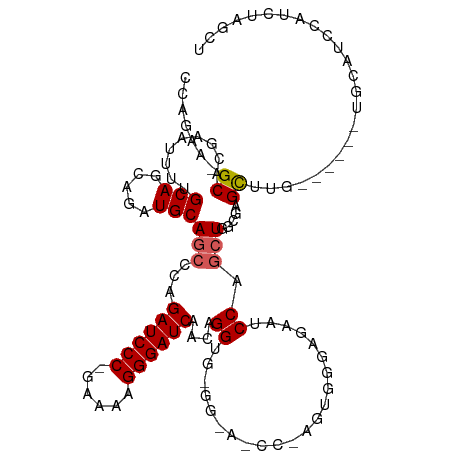
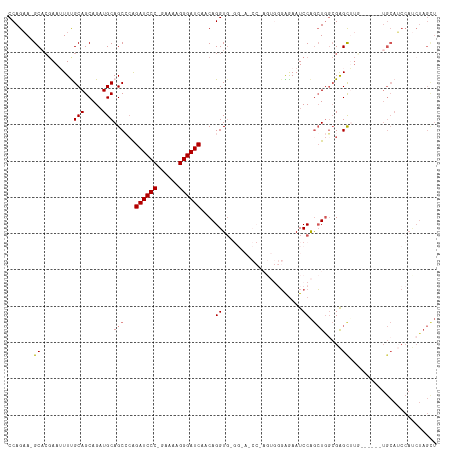
| Location | 18,729,789 – 18,729,885 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 70.85 |
| Shannon entropy | 0.49177 |
| G+C content | 0.55796 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.827859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18729789 96 - 24543557 AGCUAGAUGGAUGCA------CAUGCUCGCCAGCUGGAUUCCC--------------CACCUGUUGAUCCCUUUUC-GGGAUCUCGGCUGCAUCUGCUGCAAAAUUCGUGC-UUCUGG ..(((((.....(((------.((((..(((((.(((.....)--------------)).))...((((((.....-))))))..))).)))).))).(((.......)))-.))))) ( -30.90, z-score = -1.24, R) >droSim1.chr3L 18037498 111 - 22553184 AGCUAGAUGGAUGCA------CAUGCUCGCCAGCUGGAUUCUCCCACUCGGAUUCCGCACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGG ..(((((.....(((------(....((((.(((.(((...(((.....))).)))((((((...((((((.....-)))))).))).)))....))))))).....))))..))))) ( -39.40, z-score = -1.85, R) >droSec1.super_19 702614 79 - 1257711 -----------------------AGCUC-CCACUCGGAUUCCG--------------CACCUGUUGAUCCCUUUUC-GGGAUCUGGGCUGCAUCUGCUGCGAAAUUCGUGCUUUCUGG -----------------------.....-((((.((((((.((--------------(((((...((((((.....-)))))).)))..((....)))))).)))))).).....))) ( -23.70, z-score = -1.27, R) >droYak2.chr3L 8862964 117 + 24197627 AACUAGAUGGAUGCAGCCGGGAAAGCUCGCUAGCUGGAUUUUCCCACUGGGCUGCCCCACCUGUUGAUCCCCUUUUGGGGAUCCGCGCUGCAGCUGCUGCAAAUUUGCAGC-UUCUGG ..(((((.((.((((((.(((..(((((...((.(((......))))))))))..)))....((.(((((((....))))))).)))))))).))((((((....))))))-.))))) ( -54.30, z-score = -3.83, R) >droEre2.scaffold_4784 10567665 116 + 25762168 AACUAGGUGGCUGCAGCCGGGAAAACCCGGCAGCUGGAUUCUCCCACUCGGAUGCCCAACCUGUUGAUCCCUUUUU-GGGAUCUGCGCUGCAUCUGCUGCAAAUUUCGUGC-UGCUGG .....((..((((((((((((....))))((((((((....(((.....)))...)))....((.((((((.....-)))))).)))))))....))))))........))-..)).. ( -45.60, z-score = -2.46, R) >consensus AACUAGAUGGAUGCA______AAAGCUCGCCAGCUGGAUUCUCCCACU_GG_U_CC_CACCUGUUGAUCCCUUUUC_GGGAUCUGGGCUGCAUCUGCUGCAAAAUUCGUGC_UUCUGG .................................(((((...........................((((((......))))))...((.((....)).)).............))))) (-15.72 = -15.60 + -0.12)

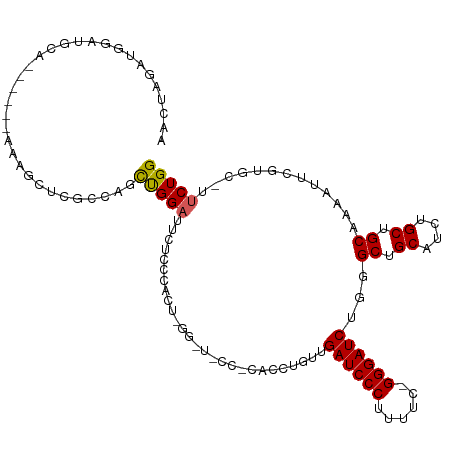
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:22 2011