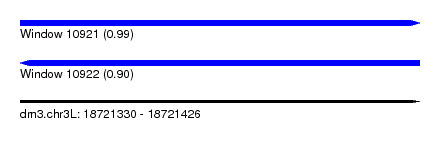
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,721,330 – 18,721,426 |
| Length | 96 |
| Max. P | 0.991065 |

| Location | 18,721,330 – 18,721,426 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.24874 |
| G+C content | 0.44525 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -19.40 |
| Energy contribution | -20.16 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.991065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
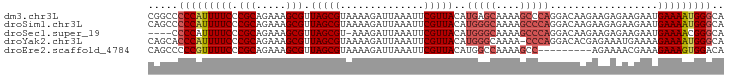
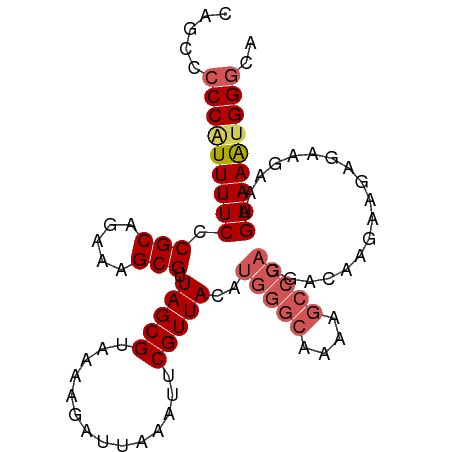
>dm3.chr3L 18721330 96 + 24543557 UGCCCAUUUUCAUUCUUCUCUUCUUGUCCUGGGCUUUUGCUCAUGUAACGAAUUUAAUCUUUUACGCUAACGCUUUCUGCGGGAAAAUGGGGGCCG ..(((((((((.........((((((.(.(((((....))))).)))).)))..................(((.....))).)))))))))..... ( -24.30, z-score = -2.05, R) >droSim1.chr3L 18027826 96 + 22553184 UGCCCAUUUUCAUUCUUCUCUUCUUGUCCUGGGCUUUUGCCCAUGUAACGAAUUUAAUCUUUUACGCUAACGCUUUCUGCGGGAAAAUGGGGGCUG ..(((((((((.........((((((.(.(((((....))))).)))).)))..................(((.....))).)))))))))..... ( -27.30, z-score = -2.94, R) >droSec1.super_19 694079 91 + 1257711 UGCCCGUUUUCAUUCUUCUCUUCUUGUCCUGGGCUUUUGCCCAUGUAACGAAUUUAAUCUUU-ACGCUAACGCUUUCUGCGGGAAAAUGGGG---- ..(((((((((..................(((((....))))).((((.((......)).))-)).....(((.....))).))))))))).---- ( -27.70, z-score = -3.79, R) >droYak2.chr3L 8854131 95 - 24197627 UGCCCAUUUUCUUUUCAUUUCUCGUGUCCUGGG-UUUUGCCCAUGUAACGAAUUUAAUCUUUUACGCUAACGCUUUCUGCGGGAAAAUGGGUGCUG .(((((((((((.........((((....((((-.....))))....))))...................(((.....)))))))))))))).... ( -29.40, z-score = -4.07, R) >droEre2.scaffold_4784 10558613 87 - 25762168 UGUCCACUUUCUUUCGUUUUCU---------GGCUUUUGGCCAUGUAACGAAUUUAAUCUUUUACGCUAACGCUUUCUGCGGGAAAACGGGGGCUG .((((..(((((((((((...(---------(((.....))))...))))))..................(((.....))))))))....)))).. ( -23.00, z-score = -2.27, R) >consensus UGCCCAUUUUCAUUCUUCUCUUCUUGUCCUGGGCUUUUGCCCAUGUAACGAAUUUAAUCUUUUACGCUAACGCUUUCUGCGGGAAAAUGGGGGCUG ..(((((((((..................(((((....)))))...........................(((.....))).)))))))))..... (-19.40 = -20.16 + 0.76)

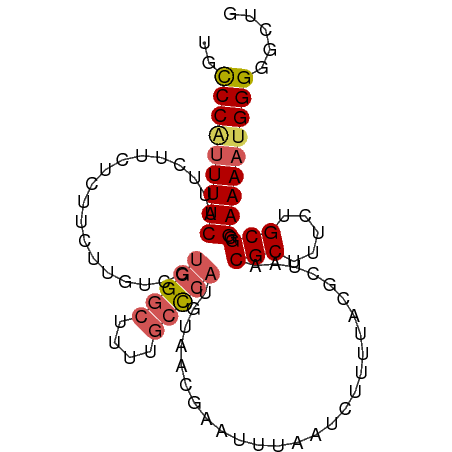
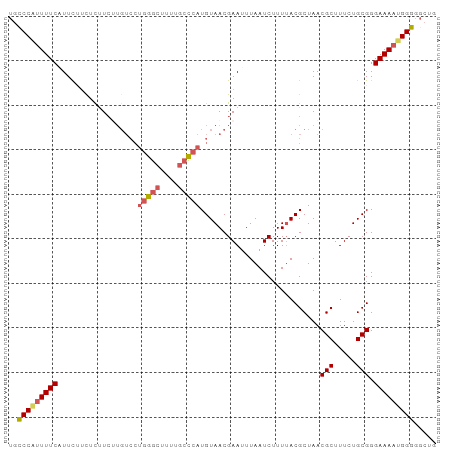
| Location | 18,721,330 – 18,721,426 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.15 |
| Shannon entropy | 0.24874 |
| G+C content | 0.44525 |
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -16.08 |
| Energy contribution | -17.20 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18721330 96 - 24543557 CGGCCCCCAUUUUCCCGCAGAAAGCGUUAGCGUAAAAGAUUAAAUUCGUUACAUGAGCAAAAGCCCAGGACAAGAAGAGAAGAAUGAAAAUGGGCA .....(((((((((.(((.....))).....................(((...((.((....)).)).)))..............))))))))).. ( -19.10, z-score = -0.83, R) >droSim1.chr3L 18027826 96 - 22553184 CAGCCCCCAUUUUCCCGCAGAAAGCGUUAGCGUAAAAGAUUAAAUUCGUUACAUGGGCAAAAGCCCAGGACAAGAAGAGAAGAAUGAAAAUGGGCA .....(((((((((.(((.....))).....................(((...(((((....))))).)))..............))))))))).. ( -26.00, z-score = -2.93, R) >droSec1.super_19 694079 91 - 1257711 ----CCCCAUUUUCCCGCAGAAAGCGUUAGCGU-AAAGAUUAAAUUCGUUACAUGGGCAAAAGCCCAGGACAAGAAGAGAAGAAUGAAAACGGGCA ----.(((.(((((..((.....))(((...((-((.((......)).)))).(((((....))))).)))..............))))).))).. ( -22.30, z-score = -2.08, R) >droYak2.chr3L 8854131 95 + 24197627 CAGCACCCAUUUUCCCGCAGAAAGCGUUAGCGUAAAAGAUUAAAUUCGUUACAUGGGCAAAA-CCCAGGACACGAGAAAUGAAAAGAAAAUGGGCA .....(((((((((.(((.....)))((..(((....((......))(((...((((.....-)))).))))))..)).......))))))))).. ( -24.80, z-score = -2.91, R) >droEre2.scaffold_4784 10558613 87 + 25762168 CAGCCCCCGUUUUCCCGCAGAAAGCGUUAGCGUAAAAGAUUAAAUUCGUUACAUGGCCAAAAGCC---------AGAAAACGAAAGAAAGUGGACA ......((((((((.(((.....)))..................((((((...((((.....)))---------)...)))))).))))))))... ( -19.90, z-score = -2.57, R) >consensus CAGCCCCCAUUUUCCCGCAGAAAGCGUUAGCGUAAAAGAUUAAAUUCGUUACAUGGGCAAAAGCCCAGGACAAGAAGAGAAGAAUGAAAAUGGGCA .....(((((((((.(((.....))).(((((..............)))))..(((((....)))))..................))))))))).. (-16.08 = -17.20 + 1.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:17 2011