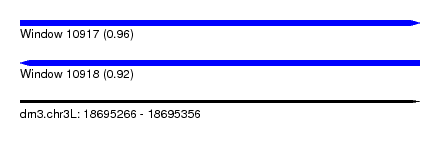
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,695,266 – 18,695,356 |
| Length | 90 |
| Max. P | 0.962456 |

| Location | 18,695,266 – 18,695,356 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.41528 |
| G+C content | 0.33629 |
| Mean single sequence MFE | -22.05 |
| Consensus MFE | -11.24 |
| Energy contribution | -11.15 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.962456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
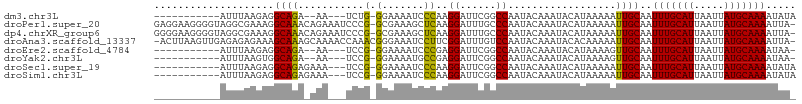
>dm3.chr3L 18695266 90 + 24543557 -----------AUUUAAGAGGCAGA--AA---UCUG-GGAAAAUCCCAAGGAUUCGGCCAAUACAAAUACAUAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUAUA -----------........(((.((--(.---((((-((.....))).))).))).)))........................(((((((.....)))))))..... ( -21.10, z-score = -2.98, R) >droPer1.super_20 1487842 105 + 1872136 GAGGAAGGGGUAGGCGAAAGGCAAACAGAAAUCCCG-GCGAAAGCUCAAGGAUUUGCCCAAUACAAAUACAUAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUUA- ..(....((((.(.(....).)......((((((.(-((....)))...))))))))))....)...................(((((((.....)))))))....- ( -27.50, z-score = -2.90, R) >dp4.chrXR_group6 11588538 105 + 13314419 GGGGAAGGGGUAGGCGAAAGGCAAACAGAAAUCCCG-GCGAAAGCUCAAGGAUUUGCCCAAUACAAAUACAUAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUUA- .......((((.(.(....).)......((((((.(-((....)))...))))))))))........................(((((((.....)))))))....- ( -27.40, z-score = -2.48, R) >droAna3.scaffold_13337 4569774 105 - 23293914 -ACUUAAGUUGAGAGAGAAAGCAAAGCAAAACCAAACGGGAAAUCCUUCGGAUUUGUCCAAUACAAAUACACAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUUA- -........................((((..((....(((....)))..))((((((.....)))))).........))))..(((((((.....)))))))....- ( -15.90, z-score = -0.64, R) >droEre2.scaffold_4784 10540380 89 - 25762168 -----------AUUUAAGAGGCAGA--AA---UCCG-GGAAAAUCCCGAGGAUUCGGCCAAUACAAAUACAUAAAAGUUGCAAUUUGCAUUAAUUAUGCAAAAUAA- -----------........(((.((--(.---((((-((.....)))).)).))).)))........................(((((((.....)))))))....- ( -22.60, z-score = -3.40, R) >droYak2.chr3L 8834992 89 - 24197627 -----------AUUUAAGUGGCAGA--AA---UCCG-GGAAAAUGCCGAGGAUUCGGCCAAUACAAAUACAUAAAAGUUGCAAUUUGCAUUAAUUAUGCAAAAUAA- -----------.......((((.((--(.---(((.-((......))..)))))).)))).......................(((((((.....)))))))....- ( -19.90, z-score = -1.79, R) >droSec1.super_19 675934 92 + 1257711 -----------AUUUAAGAGGCAGAGAAA---UCCG-GGAAAAUCCCAAGGAUUCGGCCAAUACAAAUACAUAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUAUA -----------........(((...(((.---((((-((.....)))..)))))).)))........................(((((((.....)))))))..... ( -21.00, z-score = -2.97, R) >droSim1.chr3L 18009571 92 + 22553184 -----------AUUUAAGAGGCAGAGAAA---UCCG-GGAAAAUCCCAAGGAUUCGGCCAAUACAAAUACAUAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUAUA -----------........(((...(((.---((((-((.....)))..)))))).)))........................(((((((.....)))))))..... ( -21.00, z-score = -2.97, R) >consensus ___________AUUUAAGAGGCAGA_AAA___UCCG_GGAAAAUCCCAAGGAUUCGGCCAAUACAAAUACAUAAAAAUUGCAAUUUGCAUUAAUUAUGCAAAAUAA_ ....................((((.............((......))..((......))..................))))..(((((((.....)))))))..... (-11.24 = -11.15 + -0.09)
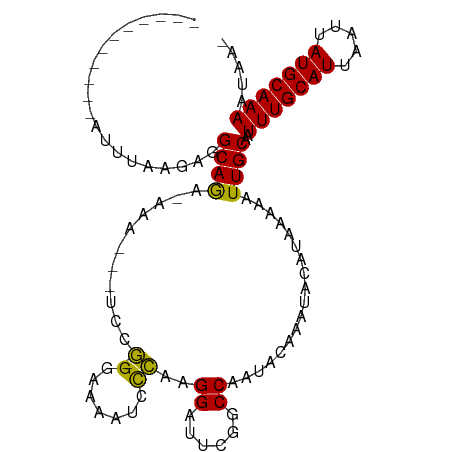
| Location | 18,695,266 – 18,695,356 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.41528 |
| G+C content | 0.33629 |
| Mean single sequence MFE | -21.61 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.34 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.918531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18695266 90 - 24543557 UAUAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCC-CAGA---UU--UCUGCCUCUUAAAU----------- ((((...(((((((..(.(((.....))).)..)))))))..))))..(((.(((..((.(((.....))-))).---.)--)).)))........----------- ( -23.20, z-score = -3.30, R) >droPer1.super_20 1487842 105 - 1872136 -UAAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGGCAAAUCCUUGAGCUUUCGC-CGGGAUUUCUGUUUGCCUUUCGCCUACCCCUUCCUC -......(((((((..(.(((.....))).)..)))))))...((...(((((((((((.(.((....))-))))))......))))))...))............. ( -20.60, z-score = -0.99, R) >dp4.chrXR_group6 11588538 105 - 13314419 -UAAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGGCAAAUCCUUGAGCUUUCGC-CGGGAUUUCUGUUUGCCUUUCGCCUACCCCUUCCCC -......(((((((..(.(((.....))).)..)))))))...((...(((((((((((.(.((....))-))))))......))))))...))............. ( -20.60, z-score = -0.88, R) >droAna3.scaffold_13337 4569774 105 + 23293914 -UAAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUGUGUAUUUGUAUUGGACAAAUCCGAAGGAUUUCCCGUUUGGUUUUGCUUUGCUUUCUCUCUCAACUUAAGU- -.......(((...(((((((((((((((.....)))).))))))))))).((((.(((((((.....)).))))).))))...)))...................- ( -19.70, z-score = -1.11, R) >droEre2.scaffold_4784 10540380 89 + 25762168 -UUAUUUUGCAUAAUUAAUGCAAAUUGCAACUUUUAUGUAUUUGUAUUGGCCGAAUCCUCGGGAUUUUCC-CGGA---UU--UCUGCCUCUUAAAU----------- -.(((..(((((((....(((.....)))....)))))))...)))..(((.(((..(.((((.....))-))).---.)--)).)))........----------- ( -23.40, z-score = -3.46, R) >droYak2.chr3L 8834992 89 + 24197627 -UUAUUUUGCAUAAUUAAUGCAAAUUGCAACUUUUAUGUAUUUGUAUUGGCCGAAUCCUCGGCAUUUUCC-CGGA---UU--UCUGCCACUUAAAU----------- -.(((..(((((((....(((.....)))....)))))))...))).((((.((((((..((......))-.)))---))--)..)))).......----------- ( -19.80, z-score = -2.27, R) >droSec1.super_19 675934 92 - 1257711 UAUAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCC-CGGA---UUUCUCUGCCUCUUAAAU----------- ((((...(((((((..(.(((.....))).)..)))))))..))))..(((.((((((..(((.....))-))))---)))....)))........----------- ( -22.80, z-score = -2.96, R) >droSim1.chr3L 18009571 92 - 22553184 UAUAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCC-CGGA---UUUCUCUGCCUCUUAAAU----------- ((((...(((((((..(.(((.....))).)..)))))))..))))..(((.((((((..(((.....))-))))---)))....)))........----------- ( -22.80, z-score = -2.96, R) >consensus _UUAUUUUGCAUAAUUAAUGCAAAUUGCAAUUUUUAUGUAUUUGUAUUGGCCGAAUCCUUGGGAUUUUCC_CGGA___UUU_UCUGCCUCUUAAAU___________ ........(((...(((((((((((.(((.......))))))))))))))..(((((.....))))).................))).................... (-11.20 = -11.34 + 0.14)

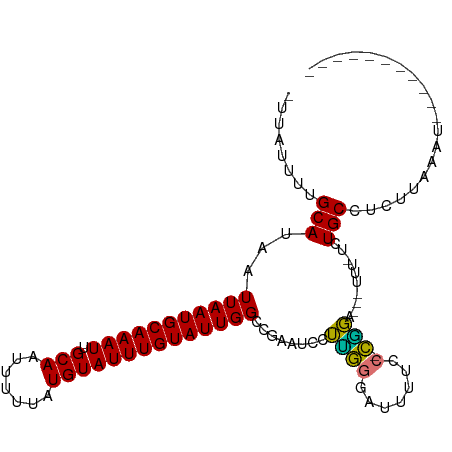
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:14 2011