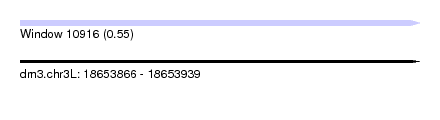
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,653,866 – 18,653,939 |
| Length | 73 |
| Max. P | 0.552868 |

| Location | 18,653,866 – 18,653,939 |
|---|---|
| Length | 73 |
| Sequences | 7 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 85.15 |
| Shannon entropy | 0.29275 |
| G+C content | 0.45411 |
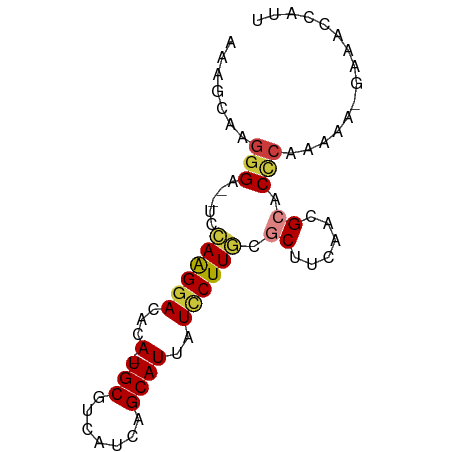
| Mean single sequence MFE | -15.01 |
| Consensus MFE | -10.06 |
| Energy contribution | -10.33 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552868 |
| Prediction | RNA |
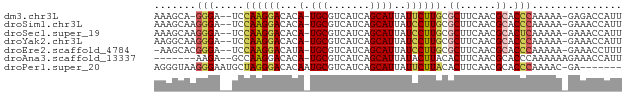
Download alignment: ClustalW | MAF
>dm3.chr3L 18653866 73 + 24543557 AAAGCA-GGGA--UCCAAGGACACA-UGCGUCAUCAGCAUUAUUCUUGCGCUUCAACGCACCCAAAAA-GAGACCAUU ......-(((.--..((((((...(-(((.......))))..)))))).((......)).))).....-......... ( -14.70, z-score = -1.10, R) >droSim1.chr3L 17965847 74 + 22553184 AAAGCAAGGGA--UCCAAGGACACA-UGCGUCAUCAGCAUUAUCCUUGCGCUUCAACGCACCCAAAAA-GAAACCAUU .......(((.--..((((((...(-(((.......))))..)))))).((......)).))).....-......... ( -17.60, z-score = -2.57, R) >droSec1.super_19 634050 74 + 1257711 AAAGCAAGGGA--UCCAAGGACACA-UGCGUCAUCAGCAUUAUCCUUGCGCUUCAACGCACUCAAAAA-GAAACCAUU .......((..--((((((((...(-(((.......))))..)))))).((......)).........-))..))... ( -16.00, z-score = -1.97, R) >droYak2.chr3L 8791927 74 - 24197627 AAGGCAAGGGA--UCCAAGGACACA-UGCGUCAUCAGCAUUAUCCUUGCGCUUCAACGCACCCAAAAA-GAAACCAUU .......(((.--..((((((...(-(((.......))))..)))))).((......)).))).....-......... ( -17.60, z-score = -1.96, R) >droEre2.scaffold_4784 10498755 73 - 25762168 -AAGCACGGGA--UCCAAGGACAUA-UGCGUCAUCAGCAUUAUCCUUGCGCUUCAACGCACCCAAAAA-GAAACCUUU -......(((.--..((((((...(-(((.......))))..)))))).((......)).))).....-......... ( -18.40, z-score = -2.59, R) >droAna3.scaffold_13337 4528388 68 - 23293914 -------AAGA--GCCAAGGACACA-UGCGUCAUCAGCAUUAUACUUACACUUCAACGCACCCAAAAAAGAAACCAUU -------....--((....(((...-...)))....))........................................ ( -4.80, z-score = 0.10, R) >droPer1.super_20 1445699 70 + 1872136 AGGGUAAGGGAAUGCUAGGGACACAAUGCGUCAUCAGCAUUAUUCUUACACUUCAACGCACCCAAAAC-GA------- ((((((((((((((((...(((.......)))...)))))).))))))).)))...............-..------- ( -16.00, z-score = -1.80, R) >consensus AAAGCAAGGGA__UCCAAGGACACA_UGCGUCAUCAGCAUUAUCCUUGCGCUUCAACGCACCCAAAAA_GAAACCAUU .......(((.....((((((.....(((.......)))...)))))).((......)).)))............... (-10.06 = -10.33 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:12 2011