
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,641,964 – 18,642,094 |
| Length | 130 |
| Max. P | 0.743485 |

| Location | 18,641,964 – 18,642,065 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 71.22 |
| Shannon entropy | 0.52615 |
| G+C content | 0.49531 |
| Mean single sequence MFE | -31.62 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.15 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.657202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
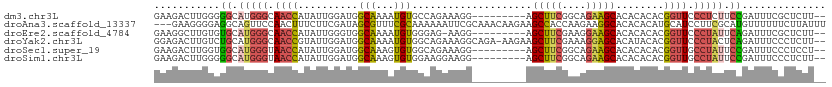
>dm3.chr3L 18641964 101 + 24543557 GAAGACUUGGGGGCAUGGGCAACCAUAUUGGAUGGCAAAAUGUGCCAGAAAGG---------AGCUUCGGCAGAAGCACACACACGGUUCCCUCUUCCGAUUUCGCUCUU-- (((((...((((((((((....))))......(((((.....)))))......---------.(((((....))))).........))))))))))).............-- ( -32.50, z-score = -1.13, R) >droAna3.scaffold_13337 4517411 109 - 23293914 ---GAAGGGGAGGCAGUUCCAACUUUCUUCGAUAGCGUUUCGCAAAAAAUUCGCAAACAAGAAGCCACCAAGAAGGCACACACAUGCAUCCUUCGCAUGUUUUUUCUUAUUU ---((((((((((.(((....))).)))))....((((((.((.........)))))).....(((........)))........))..))))).................. ( -22.30, z-score = -0.61, R) >droEre2.scaffold_4784 10487140 100 - 25762168 GAAGGCUUGUGUGCAUGGGCAACCAUAUUGGGUGGCAAAAUGUGGGAG-AAGG---------AGCUUCGAAGGAAGCACACACACGGUUCCCUAUUCAGAUUUCGCUCUU-- ....((......))((((....))))...((((((.((..((.(((((-..(.---------.(((((....))))).......)..)))))....))..))))))))..-- ( -28.60, z-score = -0.29, R) >droYak2.chr3L 8779876 109 - 24197627 GGAGACUUGUCUGCAUGGGCAACCGUAUUGGAUGGCAAAAUGUGGCAGAAAGGCAGA-AAGAAGCUUCGAAAGGAGCACAUACACGGUUCCCUACUCAGAUUUCCCUCUU-- ((((....(((((..((((.((((((.(((.....)))..((((((......))...-.....(((((....))))).)))).)))))).))))..)))))....)))).-- ( -34.70, z-score = -1.71, R) >droSec1.super_19 622087 101 + 1257711 GAAGACUUGGUGGCAUGGGUAACCAUAUUGGAUGGCAAAGUGUGGCAGAAAGG---------AGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUUCCCUCCU-- ........((.((.((((....))))((((((.(((((.(((((.(.....).---------.(((((....)))))...)))))..)))))...))))))...)).)).-- ( -34.30, z-score = -1.49, R) >droSim1.chr3L 17952261 101 + 22553184 GAAGACUUGGGGGCAUGGGUAACCAUAUUGGAUGGCAAAGUGUGGAAGGAAGG---------AGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUUCCCUCUU-- ........(((((.((((....))))((((((.(((((.(((((........(---------.(((((....))))).).)))))..)))))...))))))..)))))..-- ( -37.30, z-score = -2.58, R) >consensus GAAGACUUGGGGGCAUGGGCAACCAUAUUGGAUGGCAAAAUGUGGCAGAAAGG_________AGCUUCGAAAGAAGCACACACACGGUUCCCUAUUCCGAUUUCCCUCUU__ ...........((.(((((.((((..........(((...)))....................(((((....)))))........)))).))))).)).............. (-15.79 = -15.93 + 0.15)


| Location | 18,642,000 – 18,642,094 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.73 |
| Shannon entropy | 0.48935 |
| G+C content | 0.44993 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -9.35 |
| Energy contribution | -11.05 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
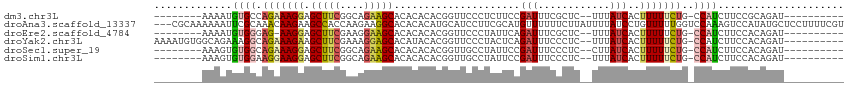
>dm3.chr3L 18642000 94 + 24543557 --------AAAAUGUGCCAGAAAGGAGCUUCGGCAGAAGCACACACACGGUUCCCUCUUCCGAUUUCGCUC--UUUAUCACUUUUUCUG-CCAUCUUCCGCAGAU---------- --------...........(((((((((.((((.((((((.(......))))...))).))))....))))--))).))......((((-(........))))).---------- ( -21.20, z-score = -1.03, R) >droAna3.scaffold_13337 4517448 112 - 23293914 ---CGCAAAAAAUUCGCAAACAAGAAGCCACCAAGAAGGCACACACAUGCAUCCUUCGCAUGUUUUUUCUUAUUUUAUCCUGUUUUUGGUCCAAAGUCCAUAUGCUCCUUUUCGU ---.(((..........(((((.((.(((........)))....((((((.......))))))..............)).))))).(((.(....).)))..))).......... ( -19.00, z-score = -0.88, R) >droEre2.scaffold_4784 10487176 93 - 25762168 --------AAAAUGUGGGAG-AAGGAGCUUCGAAGGAAGCACACACACGGUUCCCUAUUCAGAUUUCGCUC--UUUAUCACUUUUUCUG-CCAUCUUCCACAGAU---------- --------....((((((((-(.((.(((((....)))))...................((((........--............))))-)).)))))))))...---------- ( -26.65, z-score = -2.59, R) >droYak2.chr3L 8779912 102 - 24197627 AAAAUGUGGCAGAAAGGCAGAAAGAAGCUUCGAAAGGAGCACAUACACGGUUCCCUACUCAGAUUUCCCUC--UUUAUCACUUUUUCUG-CCAUCUUCCACAGAU---------- ....(((((.(((..((((((((((.(((((....)))))........((.((........))...))...--........))))))))-)).))).)))))...---------- ( -33.30, z-score = -4.38, R) >droSec1.super_19 622123 94 + 1257711 --------AAAGUGUGGCAGAAAGGAGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUUCCCUC--CUUAUCACUUUUUCUG-CCAUCUUCCACAGAU---------- --------.(((.((((((((((((.(((((....))))).......(((.........))).........--........))))))))-)))))))........---------- ( -25.80, z-score = -2.20, R) >droSim1.chr3L 17952297 94 + 22553184 --------AAAGUGUGGAAGGAAGGAGCUUCGGCAGAAGCACACACACGGUUGCCUAUUCCGAUUUCCCUC--UUUAUCACUUUUUCUG-CCAUCUUCCACAGAU---------- --------....((((((((((((...))))((((((((((.((.....))))).......(((.......--...))).....)))))-))..))))))))...---------- ( -27.30, z-score = -2.25, R) >consensus ________AAAAUGUGGCAGAAAGGAGCUUCGAAAGAAGCACACACACGGUUCCCUAUUCCGAUUUCCCUC__UUUAUCACUUUUUCUG_CCAUCUUCCACAGAU__________ ............(((((.(((.(((((((((....))))).......(((.........)))......................)))).....))).)))))............. ( -9.35 = -11.05 + 1.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:10 2011