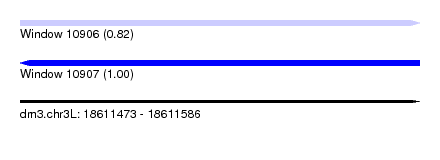
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,611,473 – 18,611,586 |
| Length | 113 |
| Max. P | 0.996310 |

| Location | 18,611,473 – 18,611,586 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Shannon entropy | 0.42130 |
| G+C content | 0.53718 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -31.91 |
| Energy contribution | -31.49 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18611473 113 + 24543557 CGCUCCCAGUACCGCAGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUU--GACGUCGGUUGUAAUUUUUU .((((((.(((((.((((((....))))).).)))))...((((((.......))))))))..(((((.......)))))..))))..(((((--(.((.....))))))))... ( -35.60, z-score = 0.14, R) >anoGam1.chr3L 13226946 108 - 41284009 GGCGCCACAUACAACAGGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCGCGC----AAGUGGACGAUUCUUU--- ((((((.(((((..((((((....))))).)..)))))..((((((.......))))))))).(((((.......)))))....)))(((..----..)))...........--- ( -42.30, z-score = -1.27, R) >droMoj3.scaffold_6680 9923560 90 - 24764193 ----------------GGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGU-------CAAAGAAAUUUUUU-- ----------------((((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))).....-------..............-- ( -32.40, z-score = -1.18, R) >droGri2.scaffold_15110 23222268 106 + 24565398 CGCGCGUUGGACAACUGGCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGA-------UAAAGUAAUAUUUU-- .(((.((..(....)..)).)))..((((..((((....(((((((.......)))))))((((.(((.......)))))))..)))).)))-------).............-- ( -35.00, z-score = -0.32, R) >droVir3.scaffold_13049 21784359 106 + 25233164 ---UCCACCUUCAACUGGCUCGUUGGUCUAGGGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGU---GGUUAAUGUAAUUAUUU--- ---.((((........((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...))---))...............--- ( -35.60, z-score = -0.98, R) >droWil1.scaffold_180949 6184520 114 - 6375548 CGCUCAACCUUCAACCGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGUGUUGAAGUAUGAAAUUAAUUUU- ...(((..(((((((((((((((..............(((((((((.......)))))).)))(((((.......))))))))))))...).)))))))..)))..........- ( -41.00, z-score = -2.32, R) >droPer1.super_20 1396271 113 + 1872136 CUCUCCAUGUUCAACUGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGGUG-GAAGUGAAUUAAAUUUUUU- ...(((((.((.....(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).)).)))-)).................- ( -36.90, z-score = -0.93, R) >dp4.chrXR_group6 11497774 113 + 13314419 CUCUCCAUGUUCAACUGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGGUG-GAAGUGAAUUAAAUUUUUU- ...(((((.((.....(((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).)).)))-)).................- ( -36.90, z-score = -0.93, R) >droEre2.scaffold_4784 10456355 112 - 25762168 CGCGUAUGCUACCGCAGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGU---GAUGUCGGUUGUAAUUUUUU ......(((.(((((((((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).....---..)).)))).)))....... ( -38.21, z-score = -0.75, R) >droYak2.chr3L 8748584 112 - 24197627 CGCUUAUGCUACCGCAGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGU---GAUGUCGGUUGUAAUUUUUU ......(((.(((((((((((((..............(((((((((.......)))))).)))(((((.......)))))))))))).....---..)).)))).)))....... ( -38.21, z-score = -0.93, R) >droSec1.super_19 591993 110 + 1257711 CGAUCCCAGUACCGCAGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGU---GAUGUCGGUUGUUUUUUU-- (((((...(((((.((((((....))))).).)))))(((..((((((.((((.(....)((((.(((.......)))))))..))))))))---)).)))))))).......-- ( -37.80, z-score = -0.77, R) >droSim1.chr3L 17922644 112 + 22553184 CGCUCCCAGUACCGCAGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGU---GAUGUCGGUUGUAAUUUUUU .((..((.(((((.((((((....))))).).)))))(((..((((((.((((.(....)((((.(((.......)))))))..))))))))---)).)))))..))........ ( -38.70, z-score = -0.94, R) >consensus CGCUCCAAGUACAACAGGCUCGUUGGUCUAGAGGUAUGAUUCUCGCUUCGGGUGCGAGAGGUCCCGGGUUCAAUUCCCGGACGAGCCCAAGU___GAUGUCGGUUAUAAUUUUU_ ................(((((((..............(((((((((.......)))))).)))(((((.......))))))))))))............................ (-31.91 = -31.49 + -0.42)
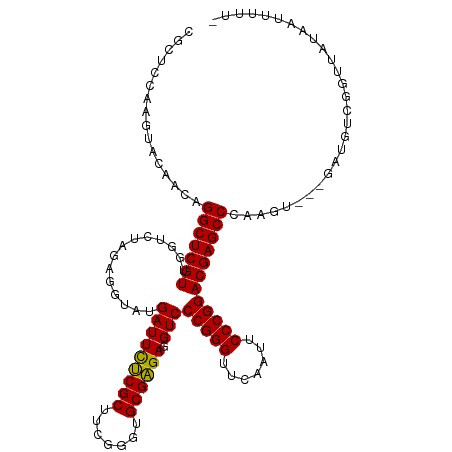
| Location | 18,611,473 – 18,611,586 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.79 |
| Shannon entropy | 0.42130 |
| G+C content | 0.53718 |
| Mean single sequence MFE | -35.29 |
| Consensus MFE | -30.02 |
| Energy contribution | -29.91 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.996310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18611473 113 - 24543557 AAAAAAUUACAACCGACGUC--AACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGCGGUACUGGGAGCG ...........((((.((..--..)..(((((((((((((.......)))))((..((((((.......)))))).))..............))))))))))))).......... ( -34.60, z-score = -1.17, R) >anoGam1.chr3L 13226946 108 + 41284009 ---AAAGAAUCGUCCACUU----GCGCGGGCUCGUCCGGGAUUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCUGUUGUAUGUGGCGCC ---.......((.((((.(----(((((((((((((((((.......)))))((..((((((.......)))))).))..............))))))))).)))).)))))).. ( -45.30, z-score = -3.78, R) >droMoj3.scaffold_6680 9923560 90 + 24764193 --AAAAAAUUUCUUUG-------ACUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCC---------------- --..............-------.....((((((((((((.......)))))......((((.......))))...................)))))))---------------- ( -26.10, z-score = -1.54, R) >droGri2.scaffold_15110 23222268 106 - 24565398 --AAAAUAUUACUUUA-------UCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGCCAGUUGUCCAACGCGCG --..............-------.....((((((((((((.......)))))......((((.......))))...................))))))).(((....)))..... ( -28.40, z-score = -0.97, R) >droVir3.scaffold_13049 21784359 106 - 25233164 ---AAAUAAUUACAUUAACC---ACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCCCUAGACCAACGAGCCAGUUGAAGGUGGA--- ---...............((---((((.((((((((((((.......)))))((..((((((.......)))))).))..............)))))))......)))))).--- ( -37.90, z-score = -3.51, R) >droWil1.scaffold_180949 6184520 114 + 6375548 -AAAAUUAAUUUCAUACUUCAACACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCGGUUGAAGGUUGAGCG -.........((((..(((((((.....((((((((((((.......)))))((..((((((.......)))))).))..............))))))).)))))))..)))).. ( -41.40, z-score = -3.82, R) >droPer1.super_20 1396271 113 - 1872136 -AAAAAAUUUAAUUCACUUC-CACCUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCAGUUGAACAUGGAGAG -................(((-((.....((((((((((((.......)))))((..((((((.......)))))).))..............))))))).(.....).))))).. ( -33.60, z-score = -2.25, R) >dp4.chrXR_group6 11497774 113 - 13314419 -AAAAAAUUUAAUUCACUUC-CACCUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCAGUUGAACAUGGAGAG -................(((-((.....((((((((((((.......)))))((..((((((.......)))))).))..............))))))).(.....).))))).. ( -33.60, z-score = -2.25, R) >droEre2.scaffold_4784 10456355 112 + 25762168 AAAAAAUUACAACCGACAUC---ACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGCGGUAGCAUACGCG ...........((((.((..---...))((((((((((((.......)))))((..((((((.......)))))).))..............)))))))..)))).((....)). ( -36.10, z-score = -2.95, R) >droYak2.chr3L 8748584 112 + 24197627 AAAAAAUUACAACCGACAUC---ACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGCGGUAGCAUAAGCG ...........((((.((..---...))((((((((((((.......)))))((..((((((.......)))))).))..............)))))))..)))).((....)). ( -36.10, z-score = -3.00, R) >droSec1.super_19 591993 110 - 1257711 --AAAAAAACAACCGACAUC---ACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGCGGUACUGGGAUCG --..........(((..(((---.(..(((((((((((((.......)))))((..((((((.......)))))).))..............))))))))).)))..)))..... ( -35.20, z-score = -2.03, R) >droSim1.chr3L 17922644 112 - 22553184 AAAAAAUUACAACCGACAUC---ACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGCGGUACUGGGAGCG ............(((..(((---.(..(((((((((((((.......)))))((..((((((.......)))))).))..............))))))))).)))..)))..... ( -35.20, z-score = -1.82, R) >consensus _AAAAAUAAUAACCGACAUC___ACUUGGGCUCGUCCGGGAAUUGAACCCGGGACCUCUCGCACCCGAAGCGAGAAUCAUACCUCUAGACCAACGAGCCUGUUGUACAUGGAGCG ............................((((((((((((.......)))))((..((((((.......)))))).))..............)))))))................ (-30.02 = -29.91 + -0.11)

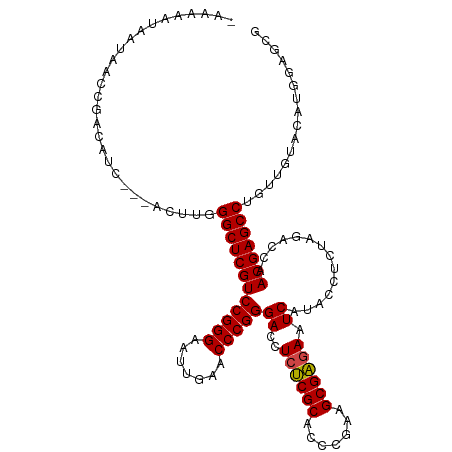
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:05 2011