| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,555,686 – 18,555,749 |
| Length | 63 |
| Max. P | 0.710169 |

| Location | 18,555,686 – 18,555,749 |
|---|---|
| Length | 63 |
| Sequences | 11 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 75.96 |
| Shannon entropy | 0.50614 |
| G+C content | 0.36198 |
| Mean single sequence MFE | -7.70 |
| Consensus MFE | -5.85 |
| Energy contribution | -5.88 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.710169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
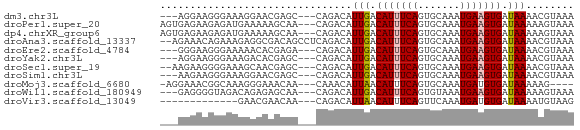
>dm3.chr3L 18555686 63 + 24543557 ---AGGAAGGGAAAGGAACGAGC---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA ---...........((......)---).........(((((((.......)))))))............ ( -7.40, z-score = -0.20, R) >droPer1.super_20 1341155 66 + 1872136 AGUGAGAAGAGAUGAAAAAGCAA---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAAAGUAAA .......................---..........(((((((.......)))))))............ ( -6.30, z-score = -0.20, R) >dp4.chrXR_group6 11443734 66 + 13314419 AGUGAGAAGAGAUGAAAAAGCAA---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAAAGUAAA .......................---..........(((((((.......)))))))............ ( -6.30, z-score = -0.20, R) >droAna3.scaffold_13337 4437707 67 - 23293914 --AGAAACAGAAAGAGGCGACAGCCUCAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA --....((.....(((((....))))).........(((((((.......))))))).......))... ( -16.10, z-score = -3.37, R) >droEre2.scaffold_4784 10404951 63 - 25762168 ---GGGAAGGGAAAAACACGAGA---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA ---.............(((....---....((((((....))))))........)))............ ( -7.19, z-score = -0.57, R) >droYak2.chr3L 8694452 63 - 24197627 ---AGGAAGGGAAAGACACGAGC---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA ---.((..((......).)...)---).........(((((((.......)))))))............ ( -7.90, z-score = -0.38, R) >droSec1.super_19 538853 64 + 1257711 --AAGAAGGGGAAAGCAACGAGC---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA --............((.....))---..........(((((((.......)))))))............ ( -6.60, z-score = 0.48, R) >droSim1.chr3L 17864534 63 + 22553184 ---AAGAAGGGAAAGGAACGAGC---CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA ---...........((......)---).........(((((((.......)))))))............ ( -7.40, z-score = -0.39, R) >droMoj3.scaffold_6680 16246870 61 + 24764193 -AGGAAACGGCAAAGGGAAACAA---CAAACAUUAACAUUUCAGUGCAAAUGAUGUGAUAAAAAG---- -.(....).(((...(....)..---....((((..(((....)))..)))).))).........---- ( -6.70, z-score = -0.96, R) >droWil1.scaffold_180949 6127443 63 - 6375548 ---GAGGGGUAGACAGAGAGCAA---CAGACAUUGACAUUUCAGUGUAAAUGAAGUGAUAAAAAGUAAA ---................((..---((.(((((((....)))))))...))..))............. ( -8.10, z-score = -1.23, R) >droVir3.scaffold_13049 21740878 53 + 25233164 -------------GAACGAACAA---CAGACAUUAACAUUUCAGUUCAAAUGAUGUGAUAAAAUGUAAG -------------..........---...(((((..(((.(((.......))).)))....)))))... ( -4.70, z-score = 0.44, R) >consensus ___GAGAAGGGAAAAAAAAGCAA___CAGACAUUGACAUUUCAGUGCAAAUGAAGUGAUAAAACGUAAA ................................(((.(((((((.......))))))).)))........ ( -5.85 = -5.88 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:03 2011