
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,531,760 – 18,531,859 |
| Length | 99 |
| Max. P | 0.777002 |

| Location | 18,531,760 – 18,531,859 |
|---|---|
| Length | 99 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.90 |
| Shannon entropy | 0.41099 |
| G+C content | 0.54638 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.777002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
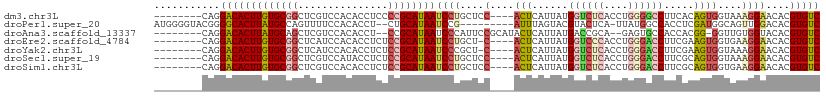
>dm3.chr3L 18531760 99 - 24543557 --------CAGGACACUUGUGCGGCUCGUCCACACCUCCCCGCAUAAUCCUGCUCC----ACUCAUUAUGGUCUCACCUGGGGCCUUCACAGUGGUAAAGGAACACGUGUC --------...(((((((((((((...((....))....))))))))((((...((----(((......((((((....)))))).....)))))...))))....))))) ( -35.70, z-score = -2.83, R) >droPer1.super_20 1317402 99 - 1872136 AUGGGGUACGGGGCACUUAUGCCAGUUUUCCACACCU--CUGCAUAAUCCG---------AUUUAGUACGUACUCA-UUAUGGCCACCUCGAUGGCAGUUGGACACGUGUC .(((((.((..((((....)))).)).))))).....--..((((..((((---------(((.....((((....-.))))((((......)))))))))))...)))). ( -24.60, z-score = 0.61, R) >droAna3.scaffold_13337 4414630 98 + 23293914 --------CAGGACACUUAUGCAGCUCGUCCACACCU--CCGCAUAAUCCCAUUCCGCAUACUCAUUAUGACCGCA--GAGUGCCACCACGG-GGUUGUGGUACACGUGUC --------...(((((....(.....)(((((((.((--(((........(((((.((....((.....))..)).--)))))......)))-)).))))).))..))))) ( -23.64, z-score = 0.41, R) >droEre2.scaffold_4784 10380946 98 + 25762168 --------CAGGACACUUGUGCGGCUCAUCCACACCUCUCCGCAUAAUCCUGCU-C----ACUCAUUAUGGUCCCACCUGGGACCUUCGAAGUGGUGAAGGAACACGUGUC --------...(((((((((((((...............))))))))((((..(-(----((.((((..((((((....)))))).....))))))))))))....))))) ( -35.96, z-score = -2.89, R) >droYak2.chr3L 8669941 98 + 24197627 --------CAGGACACUUGUGCGGCUCAUCCACACCUCUCCGCAUAAUCCCGCU-C----ACUCAUUAUGGUCUCACCUGGGACCUUCGAAGUGGUAAAGGAACACGUGUC --------...(((((((((((((...............))))))))(((.((.-(----(((......((((((....)))))).....))))))...)))....))))) ( -29.96, z-score = -1.84, R) >droSec1.super_19 515097 99 - 1257711 --------CAGGACACUUGUGCGGCUCGUCCAUACCUCUCCGCAUAAUCCUGCUCC----ACUCAUUAUGGUCUCACCUGGGACCUUCGCAGUGGUAAAGGAACACGUGUC --------...(((((((((((((...((....))....))))))))((((...((----(((......((((((....)))))).....)))))...))))....))))) ( -36.20, z-score = -3.54, R) >droSim1.chr3L 17840478 99 - 22553184 --------CAGGACACUUGUGCGGCUCGUCCACACCUCUCCGCAUAAUCCUGCUCC----ACUCAUUAUGGUCUCACCUGGGACCUUCGCAGUGGUGAAGGAACACGUGUC --------...(((((((((((((...((....))....))))))))((((.(.((----(((......((((((....)))))).....))))).).))))....))))) ( -36.40, z-score = -2.72, R) >consensus ________CAGGACACUUGUGCGGCUCGUCCACACCUCUCCGCAUAAUCCUGCUCC____ACUCAUUAUGGUCUCACCUGGGACCUUCGCAGUGGUAAAGGAACACGUGUC ...........(((((((((((((...............))))))))((((.........(((......((((((....)))))).....))).....))))....))))) (-17.62 = -18.53 + 0.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:36:02 2011