| Sequence ID | dm3.chr2L |
|---|---|
| Location | 5,642,114 – 5,642,206 |
| Length | 92 |
| Max. P | 0.999984 |

| Location | 5,642,114 – 5,642,206 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 67.94 |
| Shannon entropy | 0.70857 |
| G+C content | 0.36502 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -14.97 |
| Energy contribution | -13.51 |
| Covariance contribution | -1.46 |
| Combinations/Pair | 1.71 |
| Mean z-score | -4.45 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.73 |
| SVM RNA-class probability | 0.999984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5642114 92 + 23011544 GUCCAACUUGCCUUAGAAUAGGGGAGCUUAACUUAUGU--UUUUGAUGUUUAAGUUAAAAGCCUCUGUUCUAAGACAAUUUGAUGAUCAAAACA- ...(((.(((.(((((((((((((...((((((((...--..........))))))))...))))))))))))).))).)))............- ( -28.72, z-score = -4.32, R) >droSim1.chr2L 5435197 92 + 22036055 GUCCAACUUGCCUUAGAAUAGGGGAGCUUAACUUGUGU--UUUUAAUGUUUAAGUUAAAAGCCACUGUUCUAAGACAAUUUGAUGAUCAAAACU- ...(((.(((.((((((((((.((...((((((((...--..........))))))))...)).)))))))))).))).)))............- ( -24.42, z-score = -2.84, R) >droSec1.super_5 3719851 92 + 5866729 GUCCAACUUGCCUUAGAAUAGGGGAGCUUAACUUGUGU--UUUUAAUGUUUAAGUUAAAAGCCACUGUUCUAAGACAAUUUGAUGAUCAAAACC- ...(((.(((.((((((((((.((...((((((((...--..........))))))))...)).)))))))))).))).)))............- ( -24.42, z-score = -2.89, R) >droYak2.chr2L 8773297 92 - 22324452 GUCCCACUUGUCUUAGAAUAGGGGAGCUUAACUUAUGU--AUAUCAUGUAUAAGUUAAGAACCUUUAUUCUAAGACAAUUCGACUACAAAAUCA- (((....(((((((((((((((((..((((((((((((--((...))))))))))))))..)))))))))))))))))...)))..........- ( -36.90, z-score = -8.13, R) >droEre2.scaffold_4929 5730529 85 + 26641161 GUCCCACUUGUCUUAGAAUAGGGGAGCUUAACUUGUGU--GUGUUAUGCUUAAGUUAAGAGCCUUUAUUCUAAGUCAAUUCGACUGC-------- (((....(((.(((((((((((((..(((((((((.((--((...)))).)))))))))..))))))))))))).)))...)))...-------- ( -33.70, z-score = -5.84, R) >droAna3.scaffold_12916 11085583 94 + 16180835 GUCAAACUUGUUUUAGAAUAGGGGAGCUUAACUUAUUGGAGAGUUGAUAUUAAGUUAAGAUCCCUAAUUCUAAGAUAAUGUGACCAUUAAUGCA- ((((...((((((((((((.(((((.(((((((((.((.........)).))))))))).))))).))))))))))))..))))..........- ( -33.10, z-score = -5.78, R) >dp4.chr4_group4 3405362 93 - 6586962 GCCAAAACUAUCUUAGAUCGAAGGUUCCUGGCUUACAGGGAUUUGAUG--UAAGUUAAGAACCUCCGCUCUAAGCUAGUAUGGCUAUCAUCAACG ((((..((((.((((((.((.((((((.(((((((((.........))--))))))).)))))).)).)))))).)))).))))........... ( -36.70, z-score = -5.83, R) >droPer1.super_10 1232269 93 - 3432795 GCCAAAAUUAUCUUAGAUCGAAGGUUCCUGGCUUACAGGGAUUUGAUG--UAAGUUAAGAACCUCCGCUCUAAGCUAGUAUGGCUAUCAUCAACG ((((..((((.((((((.((.((((((.(((((((((.........))--))))))).)))))).)).)))))).)))).))))........... ( -34.40, z-score = -5.16, R) >droWil1.scaffold_180708 3938619 92 - 12563649 GUCAAACUUGUCUUAGAAUAGGGGAGCUUAACUUAUAU--UGUUUUGCUUAAAGUUAAGAGCCUCAAUUCUAAGAUAAUGCGACUAUCACCGCA- .......((((((((((((.((((..((((((((....--...........))))))))..)))).))))))))))))((((........))))- ( -31.36, z-score = -5.14, R) >droVir3.scaffold_12963 3616487 86 + 20206255 GUUUAACUUAUCUUAGAAUAGGGGAGCUUAACUUU----GUAUGAACAUUUAAGCUAAGAGCCUCAAUUCUUAGAUAAUGCGACUAACAA----- (((....((((((.(((((.((((..((((.((((----((....)))...))).))))..)))).))))).))))))...)))......----- ( -21.40, z-score = -2.85, R) >droMoj3.scaffold_6500 14019523 85 + 32352404 GUUCAACUUAUCUUAGAAUAGGGGAGCUUAACUUA----AUCUGAACACUCAAGCUAAGAGCCUCGAUUCUUCGAUAAUGCGACUAACA------ (((((..(((((..(((((.((((..((((.(((.----............))).))))..)))).)))))..))))))).))).....------ ( -18.52, z-score = -1.65, R) >droGri2.scaffold_15252 3148325 92 + 17193109 GUCAAACUUAUCUUAGAAUAGGGGAGCUUAACUUUUAUCAUAUUAGCGCUCGAGCUAAGAUCCUCAAUUCUUGGAUAAGGCGACUAGCAACU--- (((...(((((((.(((((.(((((.................(((((......)))))..))))).))))).)))))))..)))........--- ( -26.61, z-score = -2.98, R) >consensus GUCCAACUUGUCUUAGAAUAGGGGAGCUUAACUUAUGU__UUUUAAUGUUUAAGUUAAGAGCCUCUAUUCUAAGAUAAUGCGACUAUCAAAAC__ (((....(((.((.(((((.((((...(((((((.................)))))))...)))).))))).)).)))...)))........... (-14.97 = -13.51 + -1.46)

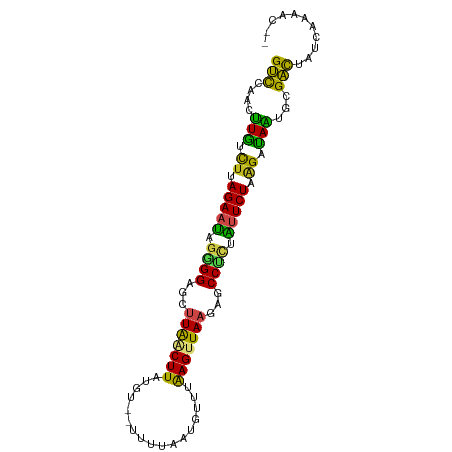
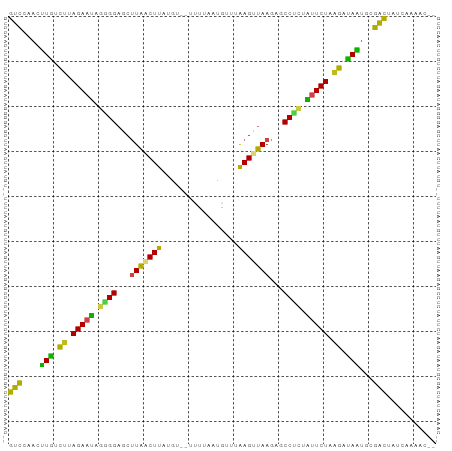
| Location | 5,642,114 – 5,642,206 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 67.94 |
| Shannon entropy | 0.70857 |
| G+C content | 0.36502 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.53 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.26 |
| SVM RNA-class probability | 0.999960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 5642114 92 - 23011544 -UGUUUUGAUCAUCAAAUUGUCUUAGAACAGAGGCUUUUAACUUAAACAUCAAAA--ACAUAAGUUAAGCUCCCCUAUUCUAAGGCAAGUUGGAC -...........((((.(((((((((((.((.((..(((((((((..........--...)))))))))..)).)).))))))))))).)))).. ( -25.62, z-score = -4.08, R) >droSim1.chr2L 5435197 92 - 22036055 -AGUUUUGAUCAUCAAAUUGUCUUAGAACAGUGGCUUUUAACUUAAACAUUAAAA--ACACAAGUUAAGCUCCCCUAUUCUAAGGCAAGUUGGAC -...........((((.(((((((((((.((.((..((((((((...........--....))))))))..)).)).))))))))))).)))).. ( -23.96, z-score = -3.38, R) >droSec1.super_5 3719851 92 - 5866729 -GGUUUUGAUCAUCAAAUUGUCUUAGAACAGUGGCUUUUAACUUAAACAUUAAAA--ACACAAGUUAAGCUCCCCUAUUCUAAGGCAAGUUGGAC -...........((((.(((((((((((.((.((..((((((((...........--....))))))))..)).)).))))))))))).)))).. ( -23.96, z-score = -3.19, R) >droYak2.chr2L 8773297 92 + 22324452 -UGAUUUUGUAGUCGAAUUGUCUUAGAAUAAAGGUUCUUAACUUAUACAUGAUAU--ACAUAAGUUAAGCUCCCCUAUUCUAAGACAAGUGGGAC -..........(((...(((((((((((((..((..((((((((((.........--..))))))))))..))..)))))))))))))....))) ( -29.90, z-score = -4.93, R) >droEre2.scaffold_4929 5730529 85 - 26641161 --------GCAGUCGAAUUGACUUAGAAUAAAGGCUCUUAACUUAAGCAUAACAC--ACACAAGUUAAGCUCCCCUAUUCUAAGACAAGUGGGAC --------...(((...(((.(((((((((..((..((((((((...........--....))))))))..))..))))))))).)))....))) ( -20.96, z-score = -2.78, R) >droAna3.scaffold_12916 11085583 94 - 16180835 -UGCAUUAAUGGUCACAUUAUCUUAGAAUUAGGGAUCUUAACUUAAUAUCAACUCUCCAAUAAGUUAAGCUCCCCUAUUCUAAAACAAGUUUGAC -.....(((((....)))))..(((((((..((((.(((((((((...............))))))))).))))..)))))))............ ( -23.46, z-score = -4.47, R) >dp4.chr4_group4 3405362 93 + 6586962 CGUUGAUGAUAGCCAUACUAGCUUAGAGCGGAGGUUCUUAACUUA--CAUCAAAUCCCUGUAAGCCAGGAACCUUCGAUCUAAGAUAGUUUUGGC ...........((((.((((.((((((.(((((((((((..((((--((.........))))))..))))))))))).)))))).))))..)))) ( -37.90, z-score = -6.04, R) >droPer1.super_10 1232269 93 + 3432795 CGUUGAUGAUAGCCAUACUAGCUUAGAGCGGAGGUUCUUAACUUA--CAUCAAAUCCCUGUAAGCCAGGAACCUUCGAUCUAAGAUAAUUUUGGC ...........((((......((((((.(((((((((((..((((--((.........))))))..))))))))))).)))))).......)))) ( -33.92, z-score = -5.09, R) >droWil1.scaffold_180708 3938619 92 + 12563649 -UGCGGUGAUAGUCGCAUUAUCUUAGAAUUGAGGCUCUUAACUUUAAGCAAAACA--AUAUAAGUUAAGCUCCCCUAUUCUAAGACAAGUUUGAC -(((((......)))))...(((((((((.(.((..((((((((...........--....))))))))..)).).))))))))).......... ( -23.06, z-score = -2.64, R) >droVir3.scaffold_12963 3616487 86 - 20206255 -----UUGUUAGUCGCAUUAUCUAAGAAUUGAGGCUCUUAGCUUAAAUGUUCAUAC----AAAGUUAAGCUCCCCUAUUCUAAGAUAAGUUAAAC -----.........((.((((((.(((((.(.((..((((((((...(((....))----)))))))))..)).).))))).))))))))..... ( -19.80, z-score = -2.84, R) >droMoj3.scaffold_6500 14019523 85 - 32352404 ------UGUUAGUCGCAUUAUCGAAGAAUCGAGGCUCUUAGCUUGAGUGUUCAGAU----UAAGUUAAGCUCCCCUAUUCUAAGAUAAGUUGAAC ------......(((..(((((..(((((.(.((..((((((((((.........)----)))))))))..)).).)))))..)))))..))).. ( -21.50, z-score = -1.93, R) >droGri2.scaffold_15252 3148325 92 - 17193109 ---AGUUGCUAGUCGCCUUAUCCAAGAAUUGAGGAUCUUAGCUCGAGCGCUAAUAUGAUAAAAGUUAAGCUCCCCUAUUCUAAGAUAAGUUUGAC ---........((((.((((((..(((((.(.(((.(((((((......(......).....))))))).))).).)))))..))))))..)))) ( -22.20, z-score = -1.70, R) >consensus __GUGUUGAUAGUCAAAUUAUCUUAGAAUAGAGGCUCUUAACUUAAACAUUAAAA__ACAUAAGUUAAGCUCCCCUAUUCUAAGACAAGUUGGAC .................((((((.(((((.(.((..((((((((.................))))))))..)).).))))).))))))....... (-13.07 = -13.53 + 0.46)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:19:35 2011