
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,515,308 – 18,515,408 |
| Length | 100 |
| Max. P | 0.768059 |

| Location | 18,515,308 – 18,515,408 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 64.23 |
| Shannon entropy | 0.73662 |
| G+C content | 0.36215 |
| Mean single sequence MFE | -22.24 |
| Consensus MFE | -6.71 |
| Energy contribution | -6.86 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.768059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18515308 100 - 24543557 ACAAAUUGAUUUGCGAGUAAGUUGAUUUCGAUUGAUUUUAAUCAAUGAGCGCUUACGAACUGCUAUUGAAAAUCAGUU--UAGGAAACCUGUGAGUAGUCCC------------- ..((((..(((((....)))))..))))..((((((....))))))((..((((((((((((....(....).)))))--).(....)..))))))..))..------------- ( -19.60, z-score = -0.75, R) >droPer1.super_20 1311506 92 - 1872136 ACAAAUUGAUUUGCGAGUAAGUUGAUUUCUAUUGAUUUGAGUCAAUAA--------GAAAUGGAAUUGCAAAUCAGAU--CAACAAACCUGUGAGCGAGCAC------------- .....(((((((((((.....((.((((((((((((....)))))).)--------))))).)).))))))))))).(--((.((....)))))........------------- ( -23.60, z-score = -2.61, R) >dp4.chrXR_group6 11413276 92 - 13314419 ACAAAUUGAUUUGCGAGUAAGUUGAUUUCUAUUGAUUUGAGUCAAUAA--------GAAAUGGAAUUGCAAAUCAGAU--CAACAAACCUGUGAGCGAGCAC------------- .....(((((((((((.....((.((((((((((((....)))))).)--------))))).)).))))))))))).(--((.((....)))))........------------- ( -23.60, z-score = -2.61, R) >droAna3.scaffold_13337 4406170 79 + 23293914 --AAAUUGAUUUGUGAGUAAGUUGAUUUUUAUUGAUUUUAAUCAAUGAACAGUU-CGCAUUCAGAUUGCAAAUCGGAG--AGAC------------------------------- --...((((((((..(....((.(((((((((((((....))))))))).))))-.)).......)..))))))))..--....------------------------------- ( -16.80, z-score = -1.38, R) >droEre2.scaffold_4784 10373354 100 + 25762168 ACAAAUUGAUUUGCGAGUAAGUUGAUUUCGAUUGAUUUUAAUCAAUGAGCGUUUGCGAACUUCCAUUGAAUAUCAGUU--UGGGAAACCCGCUAGUGGUCCC------------- .(((((((((...(((..(((((....((.((((((....))))))))((....)).)))))...)))...)))))))--))(((...(((....)))))).------------- ( -22.30, z-score = -0.82, R) >droYak2.chr3L 8662261 100 + 24197627 ACAAAUUGAUUUGUGAGUAACUUGAUUUCGAUUGAUUUUAAUCAAUGAGCGCUGGCGAACUCCUAUUGAAAAUCAGUU--UAGGAAACCUGCGAGUGCUCCC------------- ....(((((((...((((...(((....)))...)))).)))))))(((((((.(((...((((((((.....)))).--.))))....))).)))))))..------------- ( -27.50, z-score = -3.10, R) >droSec1.super_19 507636 100 - 1257711 ACAAAUUGAUUUGUGAGUAAGUUGAUUUCGAUUGAUUUUAAUCAAUGAGCGCUUGCGAACUGCUAUUGAAAAUCAGUU--UAGGAAACCUGCGAGUGGUCCC------------- ....(((((((...((((.(((((....))))).)))).)))))))((.(((((((((((((....(....).)))))--).(....)..))))))).))..------------- ( -26.40, z-score = -2.46, R) >droSim1.chr3L 17832908 100 - 22553184 ACAAAUUGAUUUGUGAGUAAGUUGAUUUCGAUUGAUUUUAAUCAAUGAGCGCUUGCGAACUGCUAUUGAAAAUCAGUU--UAGGAAACCUGCGAGUGGUCCC------------- ....(((((((...((((.(((((....))))).)))).)))))))((.(((((((((((((....(....).)))))--).(....)..))))))).))..------------- ( -26.40, z-score = -2.46, R) >droVir3.scaffold_13049 21713248 98 - 25233164 ACAAAUUGAUUUA-AACUAAGUUGAUUUUUAUUGAUUGUAUUCAAUGAGC-UCAUCUCACUCAAG---------AAGC--AGCGAAGUCAAACAG----GCCAACAAAUGGCUGU .....(((((((.-..............(((((((......)))))))((-((.(((......))---------).).--))).)))))))...(----((((.....))))).. ( -20.20, z-score = -0.85, R) >droMoj3.scaffold_6680 16206928 92 - 24764193 ACAAAUUGAUUUA-AACUAAGUUGAUUUUUAUUGAUAGAAUUCAAUGAGC-UCAGCUCAUCGCAUU------CCACAUGCAGGUCAACCAAAUGGUUACG--------------- .......(((((.-.....((((((..((((((((......)))))))).-))))))....((((.------....)))))))))((((....))))...--------------- ( -19.70, z-score = -2.07, R) >droGri2.scaffold_15110 2700317 113 + 24565398 ACAAAUUGAUUUA-AACUAAGUUGAUUUCUAUUGAUUGUCCCCAAUGAAC-UUAUCGCACUCAAGUUGUAGGCCAAGCAGAGCACAGCUAAGCAGCCGAGCAAAUAAAUGGCUAU ..((((..(((((-...)))))..)))).(((((........)))))...-.....((.......((((.(((...((..(((...)))..)).)))..)))).......))... ( -18.54, z-score = 1.10, R) >consensus ACAAAUUGAUUUGCGAGUAAGUUGAUUUCUAUUGAUUUUAAUCAAUGAGC_CUUGCGAACUGCAAUUGAAAAUCAGUU__UAGGAAACCUGCGAGUGGUCCC_____________ ..((((..(((((....)))))..))))..((((((....))))))..................................................................... ( -6.71 = -6.86 + 0.16)

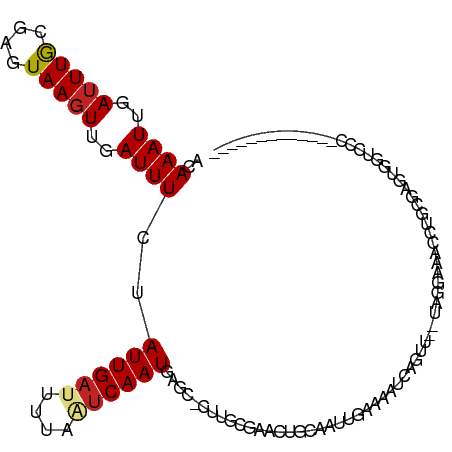

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:59 2011