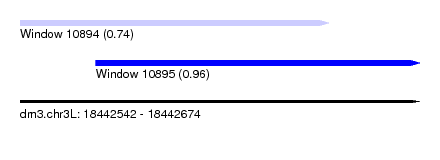
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,442,542 – 18,442,674 |
| Length | 132 |
| Max. P | 0.960347 |

| Location | 18,442,542 – 18,442,644 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Shannon entropy | 0.29545 |
| G+C content | 0.38249 |
| Mean single sequence MFE | -22.91 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.743209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
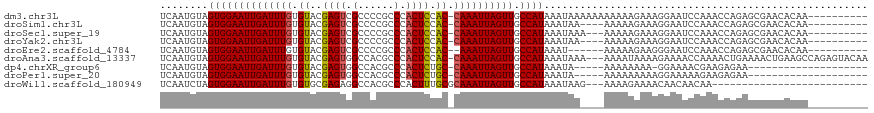
>dm3.chr3L 18442542 102 + 24543557 ---------------CUUCUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCACCA--AAUUAGUUGCCAUAAAUAAAAAAAAAA ---------------....((((..((((((((((.(((......)))(((((...((((((....))))))((...))....))))).)--)))))))))..))))............ ( -18.60, z-score = -2.11, R) >droSim1.chr3L 17776997 98 + 22553184 ---------------CUCCUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCACCA--AAUUAGUUGCCAUAAAUAAAAAA---- ---------------....((((..((((((((((.(((......)))(((((...((((((....))))))((...))....))))).)--)))))))))..))))........---- ( -18.60, z-score = -2.33, R) >droSec1.super_19 437494 99 + 1257711 ---------------CUCCUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCACCA--AAUUAGUUGCCAUAAAUAAAAAAA--- ---------------....((((..((((((((((.(((......)))(((((...((((((....))))))((...))....))))).)--)))))))))..)))).........--- ( -18.60, z-score = -2.32, R) >droYak2.chr3L 8599534 98 - 24197627 ---------------CCCUUGAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCACCA--AAUUAGUUGCCAUAAAUAAAAAA---- ---------------............(((((........)))))...((((((((((((((.((..((((.((...))..)))).))))--)))))))).))))..........---- ( -18.40, z-score = -1.96, R) >droEre2.scaffold_4784 10313343 95 - 25762168 ---------------CUCUUUACUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-A--AAUUAGUUGCCAUAAAUAAAA------ ---------------............(((((........)))))...((((((((((((((((...((((.((...))..)))))))-)--)))))))).))))........------ ( -20.80, z-score = -2.84, R) >droAna3.scaffold_13337 4336573 99 - 23293914 ---------------CUCCUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCCACCA--AAUUAGUUGCCAUAAAUAAAAAAA--- ---------------............(((((........)))))...((((((((((((((.((..((((((......)))))).))))--)))))))).))))...........--- ( -24.70, z-score = -3.37, R) >dp4.chrXR_group6 11340745 96 + 13314419 ---------------CUUUUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGCCA--AAUUAGUUGCCAUAAAUAAAA------ ---------------..(((((((...(((((........)))))...((((((((((((((.(((.((((((......)))))))))))--)))))))).)))).)))))))------ ( -28.30, z-score = -4.27, R) >droPer1.super_20 1240631 97 + 1872136 ---------------CUUUUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGCCA--AAUUAGUUGCCAUAAAUAAAAA----- ---------------..(((((((...(((((........)))))...((((((((((((((.(((.((((((......)))))))))))--)))))))).)))).))))))).----- ( -29.10, z-score = -4.52, R) >droWil1.scaffold_180949 5984836 109 - 6375548 CUUCUGAUUUGCUUUUUUGCACUCUCAAUUGAUUUUUGCUUCAAUCUAGUGGAAUUGAUUUGUGUGCGAGAGGCCACGCCCACUUUGCGC-AAAUUAGUUGCCAUAAAUA--------- ...(((((((((..((((((((.(..(((..(((.(..((.......))..))))..))).).))))))))(((...)))........))-)))))))............--------- ( -28.30, z-score = -1.93, R) >droVir3.scaffold_13049 21638188 93 + 25233164 ---------------CUACUUAUUUCAAUUGAUUCCUGCUUCAAUGUAGUGGAAUUGAUUUGUGGACGAGUGGCCACGCCUCCAUCAACA--AAUUAGUUGCCAUAAAUA--------- ---------------............(((((........)))))...(((((((((((((((.((.(.(.(((...))).)).)).)))--)))))))).)))).....--------- ( -21.80, z-score = -1.60, R) >droMoj3.scaffold_6680 16116350 95 + 24764193 ---------------CUUAUUAUUUCAAUUGAUUCCUGCUUCAAUGUAGUGGAAUUGAUUUGUGGGCGAGUGGCCACGCCUACGUCAACACAAAUUAGUUGCCAUAAAUA--------- ---------------............(((((........)))))...((((((((((((((((((((...(((...)))..))))..)))))))))))).)))).....--------- ( -25.90, z-score = -2.20, R) >droGri2.scaffold_15110 2611814 93 - 24565398 ---------------UUGCUUAUUUCAAUUGAUUCCCGCUUCAAUGUAGUGGAAUUGAUUUGUGGACGAGUGGCCACGCCUCCAUCAACA--AAUUAGUUGCCAUAAAUA--------- ---------------............(((((........)))))...(((((((((((((((.((.(.(.(((...))).)).)).)))--)))))))).)))).....--------- ( -21.80, z-score = -1.24, R) >consensus _______________CUCCUUAUUCCAAUUGAUUUUUGCUUCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCCACCA__AAUUAGUUGCCAUAAAUAAAA______ ...........................(((((........)))))...((((((((((((.(((..((........))..))).........)))))))).)))).............. (-11.37 = -11.35 + -0.02)

| Location | 18,442,567 – 18,442,674 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Shannon entropy | 0.37220 |
| G+C content | 0.39957 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.960347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18442567 107 + 24543557 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAAAAAAAAAAAGAAAGGAAUCCAAACCAGAGCGAACACAA---------- ....(((.((((((((((((((.((..((((.((...))..)))).))-)))))))))).))))...................((........)).........))).---------- ( -17.40, z-score = -1.10, R) >droSim1.chr3L 17777022 103 + 22553184 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAA----AAAAAGAAAGGAAUCCAAACCAGAGCGAACACAA---------- ....(((.((((((((((((((.((..((((.((...))..)))).))-)))))))))).))))......----.........((........)).........))).---------- ( -17.40, z-score = -0.99, R) >droSec1.super_19 437519 104 + 1257711 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAAA---AAAAAGAAAGGAAUCCAAACCAGAGCGAACACAA---------- ....(((.((((((((((((((.((..((((.((...))..)))).))-)))))))))).)))).......---.........((........)).........))).---------- ( -17.40, z-score = -1.08, R) >droYak2.chr3L 8599559 103 - 24197627 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAA----AAAAAGAAAGGAAUCCAAACCAGAGCGAACACAA---------- ....(((.((((((((((((((.((..((((.((...))..)))).))-)))))))))).))))......----.........((........)).........))).---------- ( -17.40, z-score = -0.99, R) >droEre2.scaffold_4784 10313368 100 - 25762168 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC--AAAUUAGUUGCCAUAAAU------AAAAAGAAGGGAAUCCAAACCAGAGCGAACACAA---------- ....(((.((((((((((((((((...((((.((...))..)))))))--))))))))).))))....------........((.........)).........))).---------- ( -20.30, z-score = -1.32, R) >droAna3.scaffold_13337 4336598 114 - 23293914 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCCAC-CAAAUUAGUUGCCAUAAAUAAA---AAAAUAAAAGAAAACCAAAACUGAAAACUGAAGCCAGAGUACAA ....((((((((((((((((((.((..((((((......)))))).))-)))))))))).)))).......---...........................(((....)))..)))). ( -26.00, z-score = -3.63, R) >dp4.chrXR_group6 11340770 91 + 13314419 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGC-CAAAUUAGUUGCCAUAAAUA-----AAAAAAAA-GGAAAACGAAGAGAA-------------------- ........((((((((((((((.(((.((((((......)))))))))-)))))))))).)))).....-----........-...............-------------------- ( -24.80, z-score = -3.82, R) >droPer1.super_20 1240656 92 + 1872136 UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUGGCCACGCCCACUCUGC-CAAAUUAGUUGCCAUAAAUA-----AAAAAAAAAGGAAAAAGAAGAGAA-------------------- ........((((((((((((((.(((.((((((......)))))))))-)))))))))).)))).....-----........................-------------------- ( -24.80, z-score = -4.13, R) >droWil1.scaffold_180949 5984876 89 - 6375548 UCAAUCUAGUGGAAUUGAUUUGUGUGCGAGAGGCCACGCCCACUUUGCGCAAAUUAGUUGCCAUAAAUAAG---AAAAGAAAACAACAACAA-------------------------- ........((((((((((((((((((.(((.(((...)))..))))))))))))))))).)))).......---..................-------------------------- ( -21.20, z-score = -2.06, R) >consensus UCAAUGUAGUGGAAUUGAUUUGUGUACGAGUCGCCCCGCCCACUCCAC_CAAAUUAGUUGCCAUAAAUAA____AAAAAGAAAGGAAUCCAAACCAGAGCGAACACAA__________ ........((((((((((((((.((..((((.(......).)))).)).)))))))))).))))...................................................... (-14.10 = -14.00 + -0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:54 2011