| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,440,325 – 18,440,396 |
| Length | 71 |
| Max. P | 0.939994 |

| Location | 18,440,325 – 18,440,396 |
|---|---|
| Length | 71 |
| Sequences | 12 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Shannon entropy | 0.42533 |
| G+C content | 0.52725 |
| Mean single sequence MFE | -21.94 |
| Consensus MFE | -17.61 |
| Energy contribution | -17.39 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.936684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
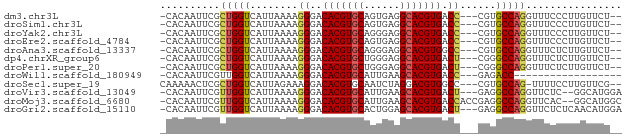
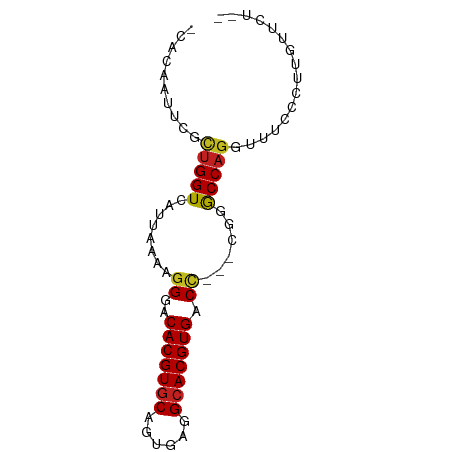
>dm3.chr3L 18440325 71 + 24543557 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCAGUGAGGCACGUGACC---CGUGCCAGGUUUCCCUUGUUCU-- -.........((((.(((.....(((.(((((((......))))))).))---))))))))..............-- ( -23.80, z-score = -1.56, R) >droSim1.chr3L 17774559 71 + 22553184 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCAGUGAGGCACGUGACC---CGUGCCAGGUUUCCCUUGUUCU-- -.........((((.(((.....(((.(((((((......))))))).))---))))))))..............-- ( -23.80, z-score = -1.56, R) >droYak2.chr3L 8597342 71 - 24197627 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCAGGGAGGCACGUGACC---CGUGCCAGGUUUCCCUUGUUCU-- -.........((((.(((.....(((.(((((((......))))))).))---))))))))..............-- ( -23.80, z-score = -1.20, R) >droEre2.scaffold_4784 10311339 71 - 25762168 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCAGUGAGGCACGUGACC---CGUGCCAGGUUUCCCUUGUUCU-- -.........((((.(((.....(((.(((((((......))))))).))---))))))))..............-- ( -23.80, z-score = -1.56, R) >droAna3.scaffold_13337 4334784 71 - 23293914 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCAGGGAGGCACGUGGCC---CGUGCCAGGUUUCUCUUGUUCU-- -.........((((.(((.....(((.(((((((......))))))).))---))))))))..............-- ( -24.20, z-score = -1.48, R) >dp4.chrXR_group6 11337458 71 + 13314419 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCUGGGAGGCACGUGACU---CGGGCCAGGUUUCUCUUGUUCU-- -.........((((((.......(((.((((((((....)))))))).))---).))))))..............-- ( -24.40, z-score = -2.14, R) >droPer1.super_20 1237465 71 + 1872136 -CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCUGGGAGGCACGUGACU---CGGGCCAGGUUUCUCUUGUUCU-- -.........((((((.......(((.((((((((....)))))))).))---).))))))..............-- ( -24.40, z-score = -2.14, R) >droWil1.scaffold_180949 395133 55 + 6375548 -CACAAUUCGUUGGUCAUUAAAAGGGACACGUGCAUUGAAGCACGUGACC---GAGACC------------------ -...........((((.......((..(((((((......))))))).))---..))))------------------ ( -16.90, z-score = -2.47, R) >droSec1.super_19 435298 71 + 1257711 CAAAAACUCGCUGGUCAUUAGAAAGGACACGUGCAAUCUAGGACGUGGCC---CGUGCCAG-UUUUCCUUGUUCG-- .........(((((.(((......((.(((((.(......).)))))..)---))))))))-)............-- ( -16.80, z-score = -0.21, R) >droVir3.scaffold_13049 21636047 71 + 25233164 -CACAAUUCGUUGGUCAUUAAAAGGGACACGUGCAUUGAAGCACGUGACU---GAGGCCAGGUUCUC--GGCAUGGA -.....(((((..(((........((.(((((((......))))))).))---((((......))))--)))))))) ( -18.40, z-score = -0.31, R) >droMoj3.scaffold_6680 16113789 74 + 24764193 -CACAAUUCGUUGGUCAUUAAAAGGGACACGUGCAUUGAAGCACGUGACCACCGAGGCCAGGUUCAC--GGCAUGGC -.....((((.(((((((.....(.....)((((......))))))))))).))))((((.((....--.)).)))) ( -23.40, z-score = -1.64, R) >droGri2.scaffold_15110 2609545 73 - 24565398 -CACAAUUCGUUGGUCAUUAAAAGGGACACGUGCACUGGAGCACGUGACU---GAGGCCAGGUUCUCUCAACAUGGA -........((((.........((((((((((((......))))))).((---(....)))..)))))))))..... ( -19.60, z-score = -0.47, R) >consensus _CACAAUUCGCUGGUCAUUAAAAGGGACACGUGCAGUGAGGCACGUGACC___CGGGCCAGGUUUCCCUUGUUCU__ ..........(((((.........((.(((((((......))))))).))......)))))................ (-17.61 = -17.39 + -0.21)

| Location | 18,440,325 – 18,440,396 |
|---|---|
| Length | 71 |
| Sequences | 12 |
| Columns | 77 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Shannon entropy | 0.42533 |
| G+C content | 0.52725 |
| Mean single sequence MFE | -21.09 |
| Consensus MFE | -12.19 |
| Energy contribution | -11.98 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
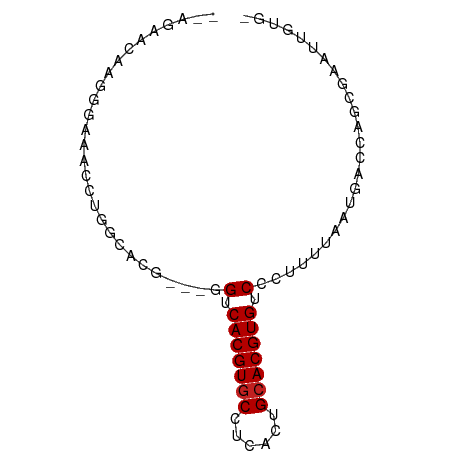
>dm3.chr3L 18440325 71 - 24543557 --AGAACAAGGGAAACCUGGCACG---GGUCACGUGCCUCACUGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((((....))(((((.(---((.(((((((......))))))).)))......)).))).....)))).- ( -25.40, z-score = -2.37, R) >droSim1.chr3L 17774559 71 - 22553184 --AGAACAAGGGAAACCUGGCACG---GGUCACGUGCCUCACUGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((((....))(((((.(---((.(((((((......))))))).)))......)).))).....)))).- ( -25.40, z-score = -2.37, R) >droYak2.chr3L 8597342 71 + 24197627 --AGAACAAGGGAAACCUGGCACG---GGUCACGUGCCUCCCUGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((((....))(((((.(---((.(((((((......))))))).)))......)).))).....)))).- ( -25.40, z-score = -2.36, R) >droEre2.scaffold_4784 10311339 71 + 25762168 --AGAACAAGGGAAACCUGGCACG---GGUCACGUGCCUCACUGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((((....))(((((.(---((.(((((((......))))))).)))......)).))).....)))).- ( -25.40, z-score = -2.37, R) >droAna3.scaffold_13337 4334784 71 + 23293914 --AGAACAAGAGAAACCUGGCACG---GGCCACGUGCCUCCCUGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((.......((((((.(---((.(((((((......))))))).)))......)).))))....)))).- ( -21.80, z-score = -2.13, R) >dp4.chrXR_group6 11337458 71 - 13314419 --AGAACAAGAGAAACCUGGCCCG---AGUCACGUGCCUCCCAGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((.......(((((..(---((.(((((((......)))))))...))).....).))))....)))).- ( -16.10, z-score = -1.44, R) >droPer1.super_20 1237465 71 - 1872136 --AGAACAAGAGAAACCUGGCCCG---AGUCACGUGCCUCCCAGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG- --...((((.......(((((..(---((.(((((((......)))))))...))).....).))))....)))).- ( -16.10, z-score = -1.44, R) >droWil1.scaffold_180949 395133 55 - 6375548 ------------------GGUCUC---GGUCACGUGCUUCAAUGCACGUGUCCCUUUUAAUGACCAACGAAUUGUG- ------------------((((..---((.(((((((......))))))).))........))))...........- ( -19.80, z-score = -3.99, R) >droSec1.super_19 435298 71 - 1257711 --CGAACAAGGAAAA-CUGGCACG---GGCCACGUCCUAGAUUGCACGUGUCCUUUCUAAUGACCAGCGAGUUUUUG --........(((((-((.((..(---(((.((((.(......).))))))))..((....))...)).))))))). ( -19.00, z-score = -1.04, R) >droVir3.scaffold_13049 21636047 71 - 25233164 UCCAUGCC--GAGAACCUGGCCUC---AGUCACGUGCUUCAAUGCACGUGUCCCUUUUAAUGACCAACGAAUUGUG- ..((((((--(......))))...---.(.(((((((......))))))).).......)))..............- ( -17.20, z-score = -1.78, R) >droMoj3.scaffold_6680 16113789 74 - 24764193 GCCAUGCC--GUGAACCUGGCCUCGGUGGUCACGUGCUUCAAUGCACGUGUCCCUUUUAAUGACCAACGAAUUGUG- (((..(((--(......))))...)))((.(((((((......))))))).)).......................- ( -22.50, z-score = -1.78, R) >droGri2.scaffold_15110 2609545 73 + 24565398 UCCAUGUUGAGAGAACCUGGCCUC---AGUCACGUGCUCCAGUGCACGUGUCCCUUUUAAUGACCAACGAAUUGUG- ....(((((.(((........)))---.(.(((((((......))))))).)............))))).......- ( -19.00, z-score = -1.62, R) >consensus __AGAACAAGGGAAACCUGGCACG___GGUCACGUGCCUCACUGCACGUGUCCCUUUUAAUGACCAGCGAAUUGUG_ .................((((......((.(((((((......)))))))...))......).)))........... (-12.19 = -11.98 + -0.21)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:53 2011