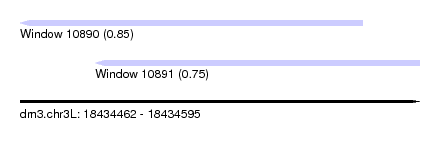
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,434,462 – 18,434,595 |
| Length | 133 |
| Max. P | 0.849925 |

| Location | 18,434,462 – 18,434,576 |
|---|---|
| Length | 114 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 90.21 |
| Shannon entropy | 0.20041 |
| G+C content | 0.36188 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -24.17 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18434462 114 - 24543557 -UUUCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA -......(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...)))))))).... ( -27.10, z-score = -1.29, R) >droSim1.chr3L 17768821 114 - 22553184 -UUUCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA -......(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...)))))))).... ( -27.10, z-score = -1.29, R) >droSec1.super_19 429487 114 - 1257711 -UUUCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA -......(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...)))))))).... ( -27.10, z-score = -1.29, R) >droYak2.chr3L 8591637 113 + 24197627 --UUCCAUUUUGCUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA --.....(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...)))))))).... ( -29.50, z-score = -1.93, R) >droEre2.scaffold_4784 10305675 115 + 25762168 UUCCCCAUUUUUCCUGCUAACCAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUGGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA ........((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))....)).)))))... ( -27.30, z-score = -1.41, R) >droAna3.scaffold_13337 4329853 114 + 23293914 -CUCCCUUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGCGCGCCUGCAUUCGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA -.....((((((((((((((((((((((((((((...(((....)))))))))...))))))).))).))).....(((((((((......)))))))))...)))))))))... ( -28.00, z-score = -1.78, R) >dp4.chrXR_group6 11330795 96 - 13314419 -GAUAUUUUUUGUUUGCUAACCAAAUAAAUGCAGCUU------------------GUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA -.......((((((((.....))))))))(((.((((------------------(..((((.....)))))))))(((((((((......))))))))).....)))....... ( -20.80, z-score = -1.31, R) >consensus _UUUCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUUUGAAUGUUCUUUUAAGCGAAAACAA .......(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).))).....(((((((((......)))))))))...)))))))).... (-24.17 = -24.87 + 0.70)

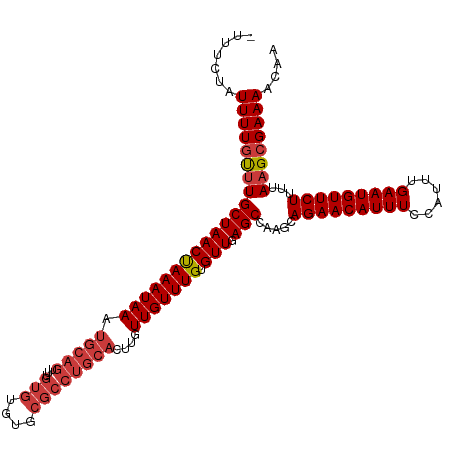
| Location | 18,434,487 – 18,434,595 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Shannon entropy | 0.32913 |
| G+C content | 0.38856 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -18.37 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.749289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
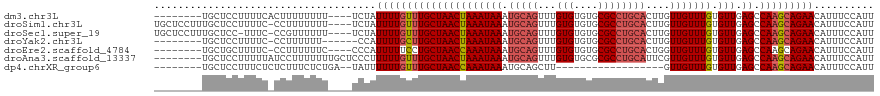
>dm3.chr3L 18434487 108 - 24543557 --------UGCUCCUUUUCACUUUUUUUU----UCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU --------.....................----....(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).......... ( -21.40, z-score = -0.75, R) >droSim1.chr3L 17768846 115 - 22553184 UGCUCCUUUGCUCCUUUUC-CCUUUUUUU----UCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU .((......))........-.........----....(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).......... ( -22.60, z-score = -1.55, R) >droSec1.super_19 429512 114 - 1257711 UGCUCCUUUGCUCC-UUUC-CCGUUUUUU----UCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU .((......))...-....-.........----....(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).......... ( -22.60, z-score = -1.29, R) >droYak2.chr3L 8591662 105 + 24197627 --------UGCUCCUUUUC-CCUUUUUU------CCAUUUUGCUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU --------...........-........------...(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).......... ( -23.80, z-score = -1.91, R) >droEre2.scaffold_4784 10305700 107 + 25762168 --------UGCUGCUUUUC-CCUUUUUUC----CCCAUUUUUCCUGCUAACCAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUGGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU --------..((((((...-.........----............(((((((((((((.(((((...(((....))))))))....))))))).))).))).))))))............ ( -20.91, z-score = -0.38, R) >droAna3.scaffold_13337 4329878 112 + 23293914 --------UGCUCCUUUUUAUCCUUUUUUUGCUCCCUUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGCGCGCCUGCAUUCGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU --------.............................(((((((((((((((((((((((((((...(((....)))))))))...))))))).))).)).))))))))).......... ( -21.80, z-score = -1.13, R) >dp4.chrXR_group6 11330820 92 - 13314419 --------UGCUCCUUUCUCUCUUUCUCUGA--UAUUUUUUGUUUGCUAACCAAAUAAAUGCAGCUU------------------GUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU --------.....................((--.((.(((((((((((((((((((((.........------------------.))))))).))).)).))))))))).)).)).... ( -14.90, z-score = -0.38, R) >consensus ________UGCUCCUUUUC_CCUUUUUUU____UCUAUUUUGUUUGCUAACUAAAUAAAUGCAGUUUGUGUGUGCGCCUGCACUUGUUGUUUGUGUUGAGCCAAGCAGAACAUUUCCAUU .....................................(((((((((((((((((((((.(((((...(((....))))))))....))))))).))).)).))))))))).......... (-18.37 = -19.07 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:51 2011