
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,399,423 – 18,399,516 |
| Length | 93 |
| Max. P | 0.867114 |

| Location | 18,399,423 – 18,399,516 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.78 |
| Shannon entropy | 0.44999 |
| G+C content | 0.48206 |
| Mean single sequence MFE | -24.21 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.54 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.867114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
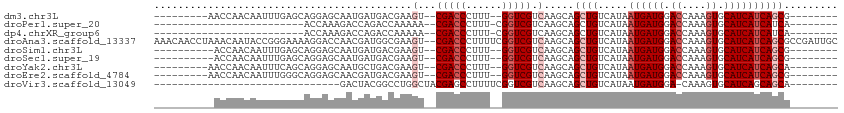
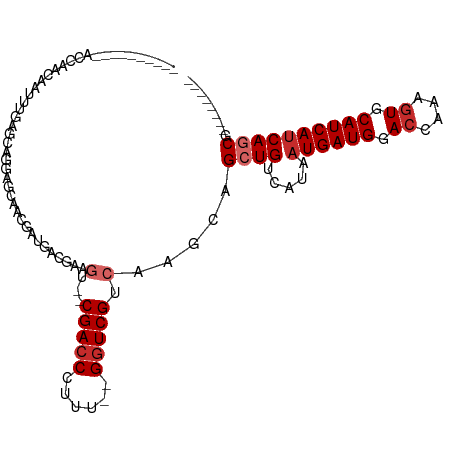
>dm3.chr3L 18399423 93 - 24543557 ---------AACCAACAAUUUGAGCAGGAGCAAUGAUGACGAAGU--CGACCCUUU--GGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCG-------- ---------....................((..(((((((((((.--.....))))--.))))))).)).((((.....((((((.((....)).)))))))))).-------- ( -23.90, z-score = -0.78, R) >droPer1.super_20 1193929 78 - 1872136 -------------------------ACCAAAGACCAGACCAAAAA--CGACCCUUU-CGGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAUCA-------- -------------------------......(((.((.......(--(((((....-.)))))).......)))))...((((((.((....)).)))))).....-------- ( -19.54, z-score = -2.61, R) >dp4.chrXR_group6 11293358 78 - 13314419 -------------------------ACCAAAGACCAGACCAAAAA--CGACCCUUU-CGGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAUCA-------- -------------------------......(((.((.......(--(((((....-.)))))).......)))))...((((((.((....)).)))))).....-------- ( -19.54, z-score = -2.61, R) >droAna3.scaffold_13337 4297750 112 + 23293914 AAACAACCUAAACAAUACCGGGAAAAGGACCAACGAUGGCGAAGU--CGACCCUUUUCGGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCGCCGAUUGC ...................((........))..(((((((...(.--(((((......))))).).....((((.....((((((.((....)).))))))))))))).)))). ( -30.00, z-score = -1.16, R) >droSim1.chr3L 17734458 92 - 22553184 ----------ACCAACAAUUUGAGCAGGAGCAAUGAUGACGAAGU--CGACCCUUU--GGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCG-------- ----------...................((..(((((((((((.--.....))))--.))))))).)).((((.....((((((.((....)).)))))))))).-------- ( -23.90, z-score = -0.74, R) >droSec1.super_19 394981 92 - 1257711 ----------ACCAACAAUUUGAGCAGGAGCAAUGAUGACGAAGU--CGACCCUUU--GGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCG-------- ----------...................((..(((((((((((.--.....))))--.))))))).)).((((.....((((((.((....)).)))))))))).-------- ( -23.90, z-score = -0.74, R) >droYak2.chr3L 8554612 93 + 24197627 ---------AACCAACAAUUUCAGCAGGAGCAAUGCUGACGAAGU--CGACCCUUU--GGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCA-------- ---------...........((((((.......))))))....(.--(((((....--))))).).....((((.....((((((.((....)).)))))))))).-------- ( -28.00, z-score = -1.98, R) >droEre2.scaffold_4784 10271125 93 + 25762168 ---------AACCAACAAUUUGGGCAGGAGCAACGAUGACGAAGU--CGACCCUUU--GGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCG-------- ---------..((((....))))((.(......)((((((((((.--.....))))--.))))))..)).((((.....((((((.((....)).)))))))))).-------- ( -25.70, z-score = -0.99, R) >droVir3.scaffold_13049 21596136 74 - 25233164 -------------------------------GACUACGGCCUGGCUACGAGCCUUUUCGGUCGUCAAGCAGCUGUCAUAAUGAUGGA-CAAAGUGCAUCAGCAGCA-------- -------------------------------....((((((.(((.....))).....))))))......(((((.....(((((.(-(...)).)))))))))).-------- ( -23.40, z-score = -1.05, R) >consensus __________ACCAACAAUUUGAGCAGGAGCAACGAUGACGAAGU__CGACCCUUU__GGUCGUCAAGCAGCUGUCAUAAUGAUGGACCAAAGUGCAUCAUCAGCG________ ...............................................(((((......))))).......((((.....((((((.((....)).))))))))))......... (-15.99 = -16.54 + 0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:46 2011