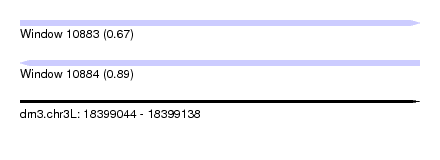
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 18,399,044 – 18,399,138 |
| Length | 94 |
| Max. P | 0.887619 |

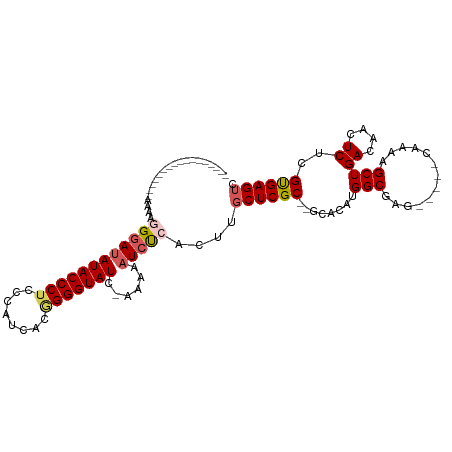
| Location | 18,399,044 – 18,399,138 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.33233 |
| G+C content | 0.49117 |
| Mean single sequence MFE | -30.66 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18399044 94 + 24543557 GACUCACGAGAGUUGUCAGCUUUUGUUUUGCUCGCCAUGUGC--GCGAGCAAGUGAUAUAUUUUGAUACCCCGUGAUGGUAGGGUAUAUCCCUACA----------------- ...(((((.(.(.((((((..(.(((((((((((((....).--))))))))..)))).)..)))))).)))))))..((((((.....)))))).----------------- ( -33.90, z-score = -3.89, R) >droSim1.chr3L 17733956 91 + 22553184 GACUCACCAGAGUUGUCAGCUUUUG-----CUCGCCAUGUGCGUGCGAGCAAGUAAGAUUUUUUGAUACCCUGUGAUGGGAGGGUAUGUCCGUUUU----------------- (((....(((((..(((....((((-----((((((......).)))))))))...))).)))))(((((((........))))))))))......----------------- ( -28.30, z-score = -1.67, R) >droSec1.super_19 394479 90 + 1257711 GACUCGCCAGAGUUGUCAGCUUUUG-----CUCGCCAUGUGCGUGCGAGCAAGUGAGAUUUUU-GAUACCCCGUGAUGGGAGGGUAUAUCCCUUUU----------------- ..(((((..((((.....))))(((-----((((((......).)))))))))))))......-....(((......)))((((.....))))...----------------- ( -28.20, z-score = -1.54, R) >droYak2.chr3L 8554199 91 - 24197627 GCCUCACGAGAGUUGUCAGCUUUUCCA--UGUCGCCAUGUGC--GCGAGCAAGUGGGAUUUUU-GAUACCCUGUUAUGGGAGGGUAUAUCCAUUUU----------------- .......((((((.....)))))).((--.(((.((((.(((--....))).)))))))...)-)(((((((.(....).))))))).........----------------- ( -25.80, z-score = -0.67, R) >droEre2.scaffold_4784 10270644 104 - 25762168 GACUCACGAGAGUUGUCAGCUUU------GGUCGCCAUGUGC--GCGAGCAAGUGGGAUUUUU-GAUACCCCGUGAUGGCCGGGUAUAUCCCUUUUCCACUAACUGAAGCAUA (((..((....)).))).((((.------.((.......(((--....)))(((((((.....-.((((((.((....)).)))))).......))))))).))..))))... ( -37.12, z-score = -2.66, R) >consensus GACUCACGAGAGUUGUCAGCUUUUG_____CUCGCCAUGUGC__GCGAGCAAGUGAGAUUUUU_GAUACCCCGUGAUGGGAGGGUAUAUCCCUUUU_________________ ..(((((.(((((.....))))).......(((((.........)))))...)))))........((((((..........)))))).......................... (-16.72 = -17.12 + 0.40)

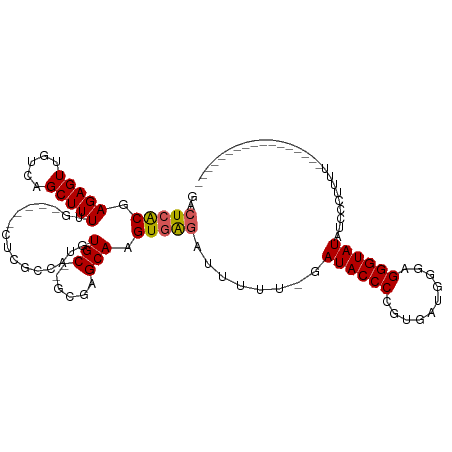
| Location | 18,399,044 – 18,399,138 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.33233 |
| G+C content | 0.49117 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -15.57 |
| Energy contribution | -16.21 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 18399044 94 - 24543557 -----------------UGUAGGGAUAUACCCUACCAUCACGGGGUAUCAAAAUAUAUCACUUGCUCGC--GCACAUGGCGAGCAAAACAAAAGCUGACAACUCUCGUGAGUC -----------------.((((((.....))))))..(((((((((.(((...........((((((((--.......)))))))).........)))..)).)))))))... ( -31.15, z-score = -4.12, R) >droSim1.chr3L 17733956 91 - 22553184 -----------------AAAACGGACAUACCCUCCCAUCACAGGGUAUCAAAAAAUCUUACUUGCUCGCACGCACAUGGCGAG-----CAAAAGCUGACAACUCUGGUGAGUC -----------------....(((.((((((((........))))))).............((((((((.((....)))))))-----)))..))))...((((....)))). ( -25.20, z-score = -2.62, R) >droSec1.super_19 394479 90 - 1257711 -----------------AAAAGGGAUAUACCCUCCCAUCACGGGGUAUC-AAAAAUCUCACUUGCUCGCACGCACAUGGCGAG-----CAAAAGCUGACAACUCUGGCGAGUC -----------------....((((((((((((........))))))).-....)))))..((((((((.((....)))))))-----))).........((((....)))). ( -26.70, z-score = -2.39, R) >droYak2.chr3L 8554199 91 + 24197627 -----------------AAAAUGGAUAUACCCUCCCAUAACAGGGUAUC-AAAAAUCCCACUUGCUCGC--GCACAUGGCGACA--UGGAAAAGCUGACAACUCUCGUGAGGC -----------------.....(((((((((((........))))))).-....))))......(((((--(..((((....))--))........((....)).)))))).. ( -22.20, z-score = -1.14, R) >droEre2.scaffold_4784 10270644 104 + 25762168 UAUGCUUCAGUUAGUGGAAAAGGGAUAUACCCGGCCAUCACGGGGUAUC-AAAAAUCCCACUUGCUCGC--GCACAUGGCGACC------AAAGCUGACAACUCUCGUGAGUC ......((((((..(((....(((((((((((.(......).)))))).-....)))))......((((--.......))))))------).))))))..((((....)))). ( -31.50, z-score = -1.58, R) >consensus _________________AAAAGGGAUAUACCCUCCCAUCACGGGGUAUC_AAAAAUCUCACUUGCUCGC__GCACAUGGCGAG_____CAAAAGCUGACAACUCUCGUGAGUC .....................((((((((((((........)))))))......)))))....((((((........(((.............)))..........)))))). (-15.57 = -16.21 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:35:45 2011